Summary
We have studied the interaction of highly purified Int protein with DNA restriction fragments from the lambda phage attachment site (attP) region. Two different DNA sequences are protected by bound Int protein against partial digestion by either pancreatic DNAase or neocarzinostatin. One Int binding site includes the 15 bp common core sequence (the crossover region for site-specific recombination) plus several bases of sequence adjoining the core in both the P and P′ arms. The second Int-protected site occurs 70 bp to the right of the common core in the P′ arm, just at the distal end of the sequence encoding Int protein. The two Int binding sites are of comparable size, 30–35 bp, but do not share any extensive sequence homology. The interaction of Int with the two sites is distinctly different, as defined by the observation that only the site in the P′ arm and not the site at the common core region is protected by Int in the face of challenge by the polyanion heparin. Restriction fragments containing DNA from the bacterial attachment site (attB) region exhibit a different pattern of interaction with Int. In the absence of heparin, a smaller (15 bp) sequence, which includes the left half of the common core region and the common core-B arm juncture, is protected against nuclease digestion by Int protein. No sequences from this region are protected by Int in the presence of heparin.
Introduction
The integration and excision reactions of bacteriophage λ constitute an extensively investigated example of site-specific recombination (for reviews see Gottesman and Weisberg, 1971; Nash, 1977). The integration of λ occurs by a reciprocal recombination between specific sites, called attachment sites, on the circularized phage chromosome and the chromosome of its host, E. coli. These sites are referred to as POP′ (attP) and BOB′ (attB). The integrated prophage is bounded by a left prophage att site, BOP′ (attL), and a right prophage att site, POB′ (attR). All four att sites are genetically distinguishable from one another (Gottesman and Weisberg, 1971). However, they all have in common a 15 bp sequence called the “common core” (“O” in the three letter designation of the att sites) within which the crossover event must occur. The DNA sequences of the four adjoining arms P, P′, B and B′ are all different from each other (Landy and Ross, 1977). At present we do not know precisely how much of each arm is involved in att site function.
The events of the λ site-specific recombination pathway are mediated by both phage and bacterial proteins (Kikuchi and Nash, 1978a, 1978b). Reconstruction of the integrative recombination reaction in vitro has led to the identification and purification of several of the required components. In this way, the phage Integrase protein (Int), whose encoding gene maps adjacent to attP, has been purified in milligram quantities to near homogeneity (Kikuchi and Nash, 1978b). In agreement with inferences drawn from earlier physiological studies, the properties of Int indicate that it is a specificity element for site-specific recombination (Kotewicz et al., 1977; Kikuchi and Nash, 1978a). Purified Int protein interacts with λ DNA to form complexes that are retained on membrane filters. The complex formation occurs more readily with λ DNA carrying attP or attL than with λ DNA carrying attB or lacking an attachment site (Kikuchi and Nash, 1978a; M. Kotewicz et al., manuscript submitted). Int therefore specifically recognizes attachment sites; sequences in the P′ arm must be required for this specific interaction. When complexes of Int and λ DNA are challenged with heparin, even larger differences in retention on filters are observed between complexes involving attP or attL as opposed to attB or no attachment site.
Int also forms heparin-resistant filter-binding complexes with several mutant or secondary attachment sites containing altered common core sequences but intact P′ arms (Kikuchi and Nash, 1978b). This observation has raised the speculation that Int may only recognize sequences in the P′ arm and not interact with sequences at the crossover locus of the core. Bound Int protein protects some DNA from the attP region from exhaustive pancreatic DNAase digestion (Kikuchi and Nash, 1978b). In the present study we have located sequences in the attachment site region that specifically interact with Int, using a technique derived from applications of the Maxam and Gilbert sequencing method (Gilbert, Maxam and Mirzabekov, 1976). This technique, called DNAase footprinting (Galas and Schmitz, 1978), identifies regions of DNA that are protected from partial pancreatic DNAase digestion by bound protein. In a restriction fragment from the attP region we find two different sequences, one including the common core region and one in the P′ arm, that are protected by Int. The interactions of Int with each of these sequences and with sequences in attB all differ from one another.
Results
Int Binding Sites on a Restriction Fragment from the attP Region
The technique of DNAase footprinting described by Galas and Schmitz (1978) utilizes a double-stranded restriction fragment labeled with 32P at one 5′ end. The labeled fragment is incubated with the purified protein and then digested with pancreatic DNAase. Conditions for digestion are selected such that each molecule is cut approximately once by the nuclease. The resultant family of end-labeled products forms a ladder on an acrylamide-urea (sequencing) gel. Presumably each base in the sequence is subject to cleavage, although some sequence specificity (preference) has been reported (Bernardi, Gaillard and Bernardi, 1975; Galas and Schmitz, 1978) and is also observed in our experiments. Ideally, the steps of the ladder represent single base increments in the sequence. The region(s) of the DNA protected from DNAase attack by the bound protein appear as interruptions in the ladder. The protected bases are identified by electrophoresing the same DNA fragment, subjected to the Maxam-Gilbert chemical sequence procedures, on adjacent lanes of the acrylamide gel. In comparing these profiles it should be recalled that pancreatic DNAase digestion yields fragments with 3′-OH termini, and that these fragments are expected to migrate more slowly than chemical sequence fragments that terminate in a 3′-PO4 (Sutcliffe and Church, 1978; McConnell, Searcy and Sutcliffe, 1978; W. Haseltine and A. D’Andrea, personal communication; A. Maxam, personal communication).
For the first set of experiments we used a cloned 240 bp restriction fragment from the attP region, with the common core sequence centrally located. This fragment is defined in the phage λ sequence by a Hinf I restriction site at −114 in the P arm and an Mnl I site at +115 in the P′ arm (see Figure 1). [Both the phage and bacterial attachment site regions are numbered with 0 as the central base of the 15 bp common core region found in each site. The P and B arm sequences are given negative numbers proceeding leftward from 0, while the P′ and B′ arm sequences are given positive numbers proceeding rightward from 0 (Landy and Ross, 1977).] The Hinf I-Mnl I fragment was labeled at the 5′ end of the bottom strand (see Experimental Procedures).
Figure 1. DNA Restriction Fragments Used in the Footprinting Experiments.
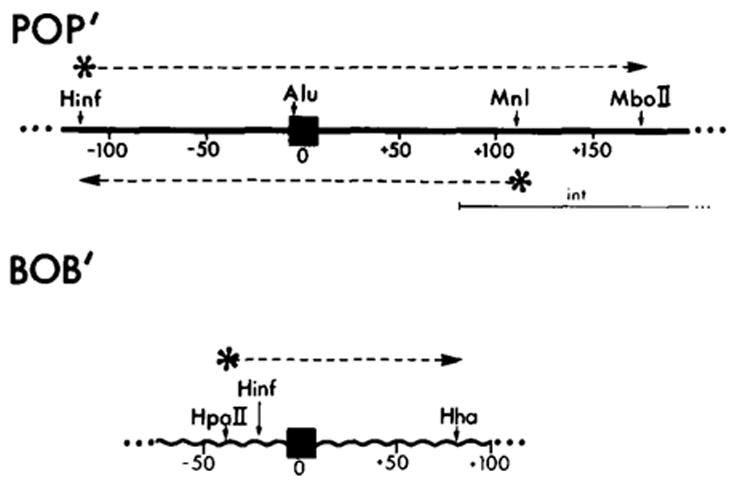
The locations of known restriction enzyme recognition sites are indicated for the POP′ (attP) and BOB′ (attB) regions studied in this work. The attachment site sequences are numbered with 0 as the central base of the 15 bp common core sequence (■); negative numbers proceed leftward from 0 into P or B arm sequences and positive numbers extend rightward from 0 into P′ or B′ arm sequences (Landy and Ross, 1977). The position of sequence encoding the carboxy terminal end of the Int protein is indicated (
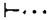 ) (see Figure 5 and Discussion). The bottom strand corresponds to the Int message. Two restriction fragments from the attP region were used: one, Hinf I (−114)-Mbo II (+173), was labeled with 32P (*) at the Hinf I site at the 5′ end of the top strand (* ‒ ‒ →); the second, Hinf I (−114)-Mnl I (+115) (← ‒ ‒ *), was labeled at the Mnl I site at the 5′ end of the bottom strand. The attB restriction fragment Hpa II (−43)-Hha I (+80) was labeled at the Hpa II site at the 5′ end of the top strand (see Experimental Procedures).
) (see Figure 5 and Discussion). The bottom strand corresponds to the Int message. Two restriction fragments from the attP region were used: one, Hinf I (−114)-Mbo II (+173), was labeled with 32P (*) at the Hinf I site at the 5′ end of the top strand (* ‒ ‒ →); the second, Hinf I (−114)-Mnl I (+115) (← ‒ ‒ *), was labeled at the Mnl I site at the 5′ end of the bottom strand. The attB restriction fragment Hpa II (−43)-Hha I (+80) was labeled at the Hpa II site at the 5′ end of the top strand (see Experimental Procedures).
DNAase footprint experiments were carried out with this attP restriction fragment under four different conditions: high and low concentrations of purified Int protein, each with and without heparin challenge (see Figure 2). When the binding reaction is carried out at a low concentration of Int and no heparin challenge is used prior to DNAase digestion, two regions are protected from DNAase attack; that is, two distinct interruptions in the ladder are observed when compared with a control digested in the absence of Int (Figure 2A, lanes 3 and 5; Figure 2B, lanes 3, 4 and 5). One of the Int-protected regions includes the 15 bp common core sequence plus several base pairs of adjoining sequence in both the P and P′ arms. The limits of protection extend from approximately −17 to +16 (see Figure 5). The other Int-protected region is somewhat distant from the crossover region for recombination (the common core), and for this reason was quite unexpected. It includes sequence from positions +48 to +85. When these Int-DNA complexes are challenged with heparin prior to digestion of the fragment with DNAase (Figure 2A, lane 6), both complexes are dissociated and no protection of the fragment is observed.
Figure 2. Pancreatic DNAase Footprints of Int on the Bottom Strand of a Restriction Fragment from the attP Region.
The fragment Hinf I (−114)-Mnl I ( + 115) is labeled with 32P at the Mnl I end (see Figure 1). Lanes 1 and 2 in each panel contain DNA sequence markers. A + G and C + T, prepared by the method of Maxam and Gilbert(see Experimental Procedures). See Figure 1 legend for numbering of sequences. The sequence is presented in Figure 5 and in Landy and Ross (1977). In (A), the DNA in samples 3 and 4 was digested with DNAase in the absence of Int, either with (lane 4) or without (lane 3) the prior addition of 5 μg/ml heparin. DNA samples in lanes 5–8 were incubated with either a low concentration of Int (1.25 μg/ml, lanes 5 and 6) or a high concentration of Int (10 μg/ml, lanes 7 and 8). Int-DNA complexes in samples 6 and 8 were also challenged with 5 μg/ml heparin prior to DNAase digestion (see Experimental Procedures). In (B), where the samples were electrophoresed for longer times to resolve the common core region better, lane 3 contains a control sample(DNAase digested in the absence of Int) while samples 4 and 5 were incubated with 1.25 (4) or 2.5 μg/ml (5) of Int (low Int) prior to DNAase digestion. No heparin was used in any of the samples in (B). DNA sequences protected from DNAase digestion are bracketed and the sequence coordinates of the boundaries of the protection are indicated beside the brackets. Two coordinates for a protection boundary reflect uncertainty as to the precise position of the boundary (see Results and Figure 5). Pancreatic DNAase digestion products have slightly different electrophoretic mobility than sequence markers (see Results).
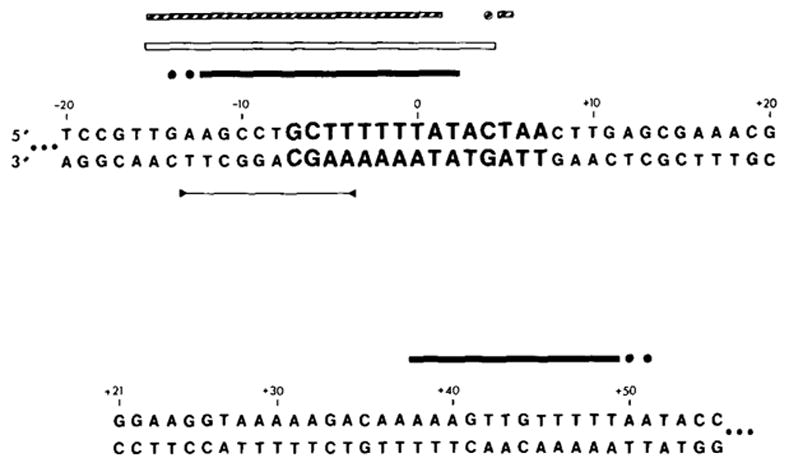
Figure 5. Two Sequences from the attP Region Protected by Int from Digestion with Either Neocarzinostatin or Pancreatic DNAase.
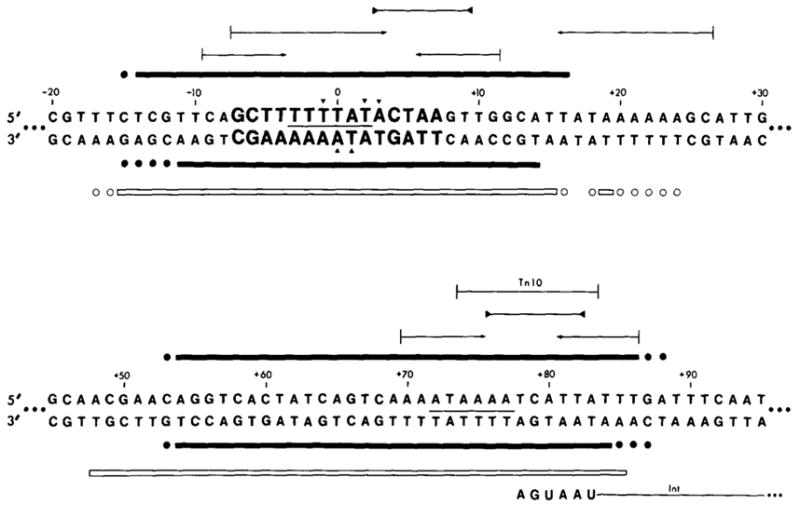
The top sequence includes the common core protected region; letters in bold type are the 15 bases of the common core sequence. The bottom sequence includes the protected region in the P′ arm. See Figure 1 legend for sequence numbering. Solid bars (■) indicate sequences protected from NCS digestion; open bars (□) indicate sequences protected from pancreatic DNAase digestion. Bars are drawn above the sequence when the data were obtained from a fragment labeled in the top strand, and below the sequence when the fragment used was labeled in the bottom strand. A base is designated as being protected from pancreatic DNAase if its 5′ phosphodiester linkage is protected. (●) or (○) indicate bases whose extent of protection from NCS or DNAase digestion is not known, due to failure of the nuclease to cut the base in an unprotected control sample (see Results). Two tandem stop codons (5′...UAAUGA...) which define the carboxy terminal end of the Int gene are indicated on the bottom strand (Int message strand; see Discussion). A 10 base sequence which is also found as part of an inverted repeat structure at the ends of Tn10 is indicated in the P′ arm protected region (Kleckner, 1979). A sequence homology between the common core and P′ arm protected regions is indicated with a line (—) between the two strands of the sequences. (
 ) Inverted repeat; (
) Inverted repeat; (
 ) palindromic sequences. Less well protected bases in NCS experiments with the top strand (▼); or the bottom strand (▲) (see Results).
) palindromic sequences. Less well protected bases in NCS experiments with the top strand (▼); or the bottom strand (▲) (see Results).
When an 8 fold higher concentration of Int is used in the binding reaction the results are quite different. In the absence of heparin challenge of the Int-DNA complex, the extent of DNAase digestion of the fragment is greatly reduced, as if the entire fragment is protected by Int binding (Figure 2A, lane 7). This effect appears to be due to a nonspecific interaction of Int protein with DNA, as it has been observed with several DNA fragments, including some which do not contain an att site and show no specific binding sites under any conditions tested (data not shown). This effect is not due to an interaction between Int and pancreatic DNAase since the nonspecific protection of DNA fragments by Int protein has also been observed in footprint experiments with other nucleases (see below and Figures 3, 4 and 6). These nonspecific Int-DNA complexes are dissociated in the presence of heparin (Figure 2A, lane 8).
Figure 3. Neocarzinostatin Footprints of Int on the Bottom Strand of a Restriction Fragment from the attP Region.
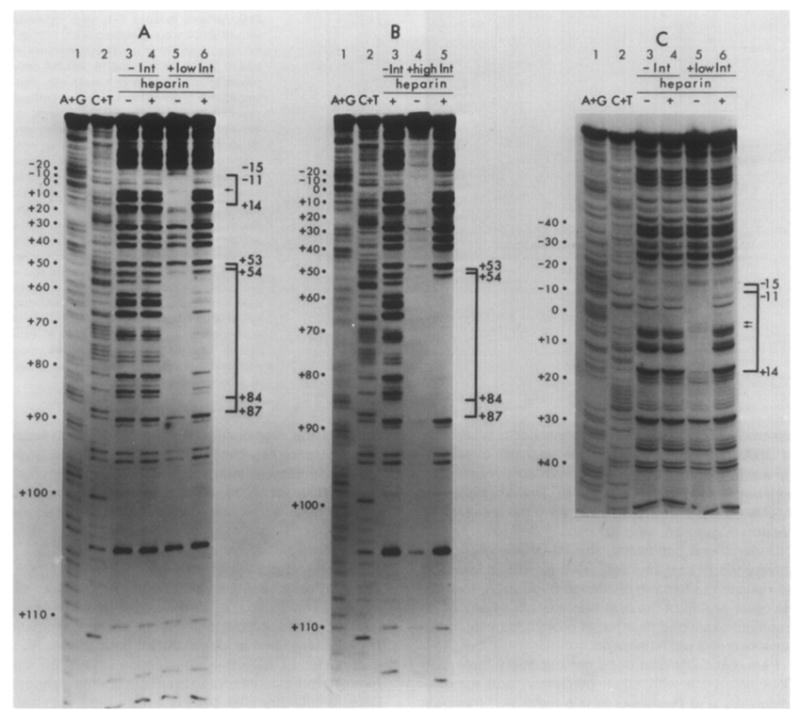
The restriction fragment Hinf I-Mnl I is the same as that used in Figure 2 (see Figure 1 and Experimental Procedures). Chemical sequence markers, prepared by the method of Maxam and Gilbert, are found in lanes 1 and 2 in each panel. Numbering of the sequence is as described in the legend to Figure 1. Samples in (A) and (C) are identical; the gel in (C) was electrophoresed for a longer time to resolve the common core region better. Samples in lanes 3 and 4 of (A) and (C) were digested with NCS in the absence of Int, and either with (lanes 4) or without (lanes 3) the prior addition of 5 μg/ml heparin. Samples in lanes 5 and 6 were incubated with 1.25 μg/ml Int (low Int), then either challenged with heparin (lanes 6) or not challenged (lanes 5) prior to NCS digestion. (B) illustrates NCS digestion profiles of samples incubated with a high concentration of Int (10 μg/ml), with (lane 5) or without (lane 4) subsequent challenge with heparin. Brackets identify the regions of sequence which are protected by Int from NCS digestion, and the sequence coordinates indicate the boundaries of protection. See Figure 5 for sequence detail. Arrows (←) within brackets indicate less well protected bases in the common core sequence (see Results).
Figure 4. Neocarzinostatin Footprints of Int on the Top Strand of a Restriction Fragment from the attP Region.
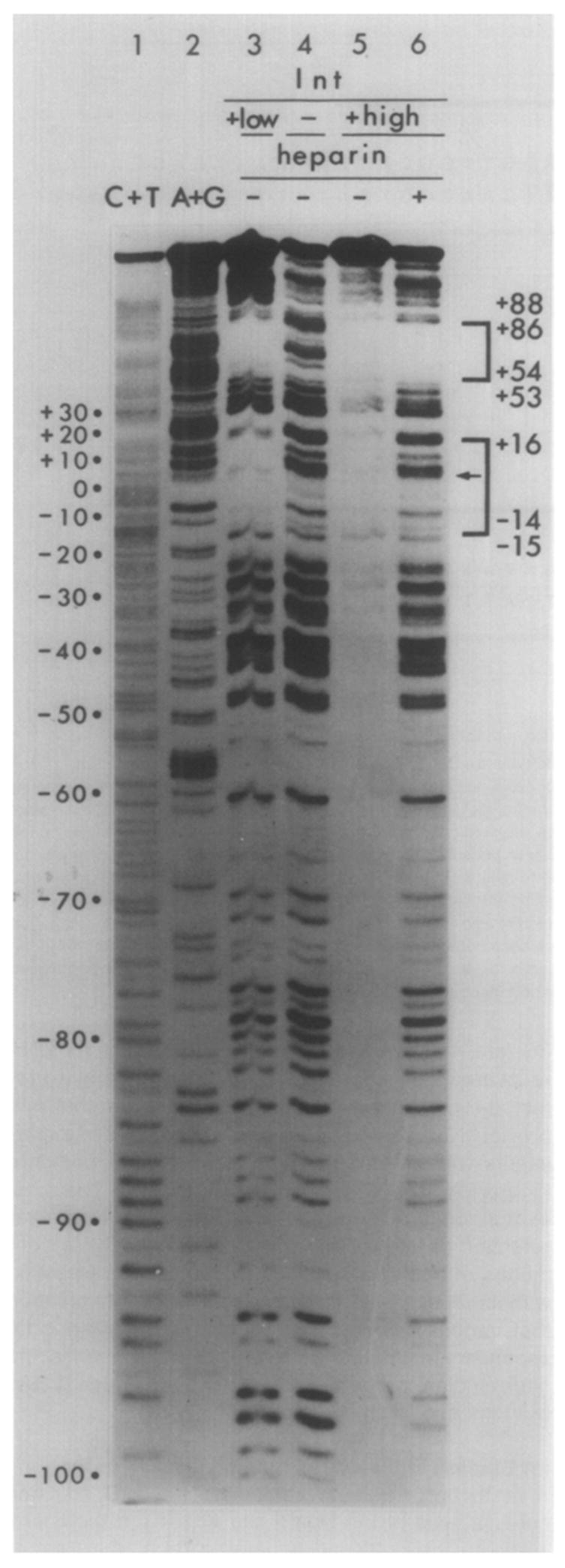
The fragment used was Hinf I (−114)-Mbo II (+173), labeled at the Hinf I end (see Figure 1). C + T and A + G chemical sequence markers (see legend to Figure 2) are in lanes 1 and 2. A control sample, digested with NCS in the absence of Int and heparin, is in lane 4. (Heparin does not influence the extent or specificity of NCS cleavage; see Figure 3A, lanes 3 and 4.) In lane 3, the DNA sample was incubated with 1.25 μg/ml Int (low Int) and not challenged with heparin prior to NCS digestion. DNA in samples 5 and 6 was incubated with a high concentration of Int (10 μg/ml), with (lane 6) or without (lane 5) challenge by heparin prior to NCS digestion. Following the digestion, samples 5 and 6 were passed through nitrocellulose filters, and the DNA retained on the filters was eluted off and applied to the gel (see Experimental Procedures). Brackets designate sequence protected from NCS cleavage by Int (see Figure 5). The boundaries of protection were determined from a gel electrophoresed for a longer time (not shown). The arrow (←) in the lower bracket indicates less well protected bases in the common core protected region.
Figure 6. Pancreatic DNAase (A), Neocarzinostatin (B) and Micrococcal Nuclease (C) Footprints of Int on the Top Strand of a Restriction Fragment from the attB Region.
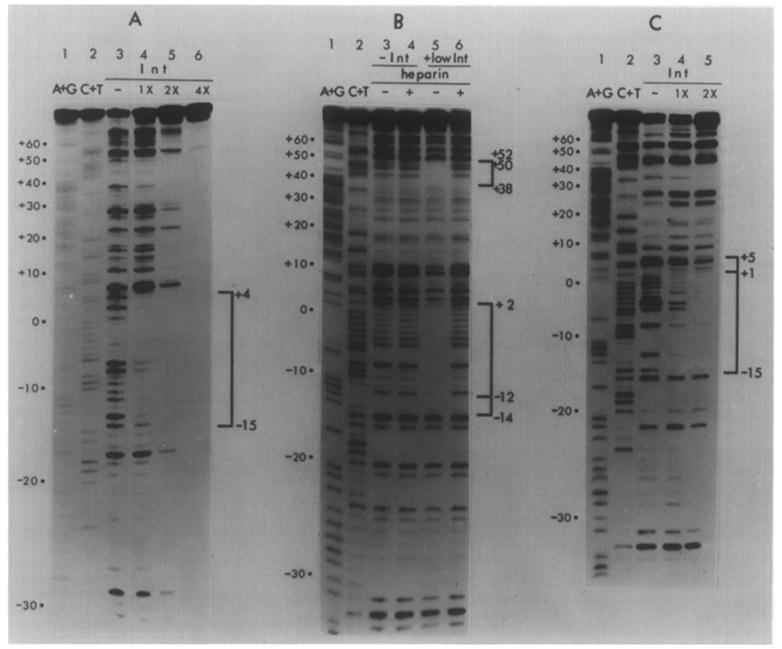
The restriction fragment Hpa II (−43)-Hha I (+80) is labeled at the Hpa II end (see Figure 1). Chemical sequence markers, as described in the legend to Figure 2 and Experimental Procedures, are in lanes 1 and 2. In (A) and (C), DNA samples were incubated without Int (lane 3) or with varying concentrations of Int [1.25 μg/ml (1X) (lane 4), 2.5 μg/ml (2X) (lane 5) or 5 μg/ml (4X) (lane 6, panel A)] prior to digestion with the specified nuclease. No heparin was used in these experiments. In (B), DNA samples were incubated in the absence of Int (lanes 3 and 4) or in the presence of 1.25 μg/ml (low) Int (lanes 5 and 6). Incubated samples in lanes 4 and 6 were challenged with heparin prior to NCS digestion. Brackets identify regions protected by Int from digestion. Protected sequences are illustrated in Figure 7. The boundaries of the region at +38 to + 50–52 were determined from a second gel (not shown).
Under these conditions, the Int-DNA complex involving the P′ arm site from +48 to +85 is found to be resistant to heparin challenge (Figure 2A, lane 8). No other region of the attP fragment, or of any other fragment examined thus far, is protected by Int in the presence of 5 μg/ml heparin.
Pancreatic DNAase does not cut every base in a DNA sequence with equal efficiency, and approximately 20% of the bases in this restriction fragment are either not cut at all or are cut with minimal efficiency in the absence of Int (see Figure 2). Precise definition of the boundaries of the protected regions is therefore limited by the digestion pattern of the control lane in these regions. These considerations are reflected in the summary of the protection data (Figure 5). The right boundary of the common core protected region is of interest due to its proximity to one arm of a large inverted repeat (see Figure 5) that is of possible significance in the recombination reaction. Footprint experiments with a different nuclease provided additional information about this region (see below). No evidence of specific protection of any other regions between the Mnl I and Hinf I sites was observed.
Neocarzinostatin Footprint Experiments
The attP restriction fragment used in the above experiments was also analyzed in a footprint experiment in which neocarzinostatin rather than pancreatic DNAase was used to cleave the Int-bound DNA. Neocarzinostatin (NCS) is a polypeptide of 10,700 daltons; it makes single-strand breaks in double-stranded DNA. Single-stranded DNA is a very poor substrate (Hatayama et al., 1978). The cleavage is specific for A and T residues; Ts are cut with relatively high although variable efficiency, while only some As are cut, with somewhat lower but also variable efficiency (D’Andrea and Haseltine, 1978; Hatayama et al., 1978). The digestion products terminate in a 3′-PO4 and thus co-migrate with fragments of the same length in base pairs generated by the chemical sequence method (D’Andrea and Haseltine, 1978).β-mercaptoethanol is required for digestion with NCS, but in contrast to pancreatic DNAase there is no divalent cation requirement. Thus the protection experiments with Int can be carried out in the presence of 1 mM EDTA, as was the case in previous filter-binding studies (Kikuchi and Nash, 1978a; see Experimental Procedures).
The results obtained with NCS partial digestion of the Int-bound fragment are in very good agreement with those generated by pancreatic DNAase for each of the four conditions analyzed (see Figures 3 and 5). The following additional points are noted.
Roughly in the center of the protected core region are two bases, the T at +1 and the A at 0, which appear to be less well protected than those bases defining the rest of the region (Figure 3C). The same phenomenon is also observed on the gel in Figure 3A, but was not observed with pancreatic nuclease (Figure 2B). This could be a reflection of the small size of the NCS molecule; the DNA conformation at the site of interaction with Int may permit access to these bases by the small NCS molecule, but not by the somewhat larger (30,000 daltons) pancreatic nuclease molecule. A similar observation has been made for the other DNA strand in this region (see below).
The precise definition of boundaries of the regions protected from NCS digestion is subject to the same limitations noted for DNAase digestion, in each case resulting from failure of the nuclease to cut every base in the unprotected sequence. The P′ arm site protected in NCS experiments is slightly shorter than that observed in DNAase experiments, perhaps due to greater accessibility of the boundary regions to the smaller NCS molecule. The NCS boundary definitions are summarized in Figure 5.
The question concerning protection of the rightmost element of the large inverted repeat in the common core region is clarified by NCS experiments (lane 5 of Figure 3C), since the right common core boundary is more precisely defined here than in the pancreatic DNAase experiment. Base +14 (T) is (partially) protected, while base +15 as well as bases +16 to +26 are protected very slightly, if at all (see Figure 5).
Analysis of Int Binding Using the Complementary Strand
To examine the boundaries of Int protection of the other strand (the top strand) of the attP region, the Hinf I site at −114 in the P arm was labeled. The 288 bp restriction fragment used in these experiments, Hinf I (−114)-Mbo II (+173) (see Figure 1), was obtained directly from phage DNA (as opposed to being a cloned restriction fragment; see Experimental Procedures).
For each of the conditions analyzed the results obtained with NCS partial digestion of this fragment (Figure 4) are in good agreement with those obtained from the bottom DNA strand (see Figure 5). Precise definition of the boundaries of the protected regions was obtained by electrophoresis on 29 inch sequencing gels (data not shown).
As described above for NCS profiles of the bottom strand in the core region (Figures 3A and 3C), the top strand also contains centrally located bases that are less well protected: the T at −1 and the TA at +2 and + 3 (Figure 4, lane 3; and gel not shown). The TA at 0 and +1 are well protected (recall that on the bottom strand the less well protected bases are at 0 and +1; see Figure 5).
An additional feature of the experiments carried out with this Hint l-Mbo II restriction fragment provides a direct correlation between these Int protection experiments and the results from previous filter-binding studies of Int complexes with intact phage DNAs (Kikuchi and Nash, 1978a). Int (at the high concentration) was incubated with the end-labeled restriction fragment, with or without heparin challenge, under the conditions used in the filter-binding studies. Following partial NCS digestion and filtration, the bound Int-DNA complexes were eluted from the nitrocellulose filters and subjected to gel electrophoresis. In the sample not challenged with heparin the DNA recovered from the filters shows the same nonspecific Int protection observed previously at the high Int concentration (Figure 4, lane 5; compare Figure 2A, lane 7 and Figure 3B, lane 4). In the heparin-challenged sample the DNA recovered from the filter shows only the single protected region in the P′ arm, the same region that was previously observed under these conditions in experiments without filtration (Figure 4, lane 6; compare Figure 2A, lane 8 and Figure 3B, lane 5). The most straightforward interpretation of this result is that filter retention of Int-att DNA complexes under these conditions of heparin challenge is due to Int interaction with the P′ arm site. This accounts for the observation that various common core sequence alterations (in secondary or mutant att sites) do not influence the filter-binding properties of the whole phage DNAs (Kikuchi and Nash, 1978b).
Int Binding in the attB Common Core Region
A restriction fragment containing the attB common core, labeled at the Hpa II site at −43 in the B arm and with the right end generated by a Hha I cut at +80 in the B′ arm (see Figure 1), was used in a series of footprint experiments. Three different nucleases (pancreatic DNAase, neocarzinostatin or micrococcal nuclease) were used in different experiments to generate partial digestion ladders of Int-bound attB fragment (see Figure 6). When samples are digested with pancreatic DNAase in the absence of heparin challenge (Figure 6A), higher concentrations of Int (lanes 5 and 6) result in increasing degrees of nonspecific binding to the entire attB fragment, as was observed with each strand of attP (Figures 2, 3 and 4) and with non-att fragments (data not shown). At a lower Int concentration (lane 4) specific protection is observed in the attB core region extending from −15 to +4. This region is about half the size of each of the two regions protected on attP fragments.
In Figure 6B, where samples were digested with NCS, a comparable region of protection extending from − 14 to + 2 is observed at a low Int concentration with no heparin challenge (lane 5). No heparin-resistant binding was observed using this Int concentration (lane 6) or a higher Int concentration (data not shown). The NCS profile also shows a second region of apparent protection involving bases +38 to +52 in the B′ arm, a region also about 15 bp in length. (This region was resolved on a gel not shown.) This second site is not discernible with either pancreatic nuclease (Figure 6A) or micrococcal nuclease (Figure 6C), due possibly to either the failure of these nucleases to cut this region effectively in the control samples or the slightly different binding conditions used.
A third nuclease, micrococcal nuclease, was used to examine the attB-Int interaction (Figure 6C). The products of micrococcal nuclease digestion terminate in 3′-PO4 and thus co-migrate with the chemical sequence markers. Partial protection of 10 micrococcal nuclease sites is shown in lanes 4 and 5 of Figure 6C. These ten sites cover the region from −15 to 0 and, with less certainty, the +1 to +5 region.
Thus the results of three different types of footprint experiments (summarized in Figure 7) define an approximately 15 bp region of protection in attB covering the left side of the common core region and 7 or 8 bases to the left in the B arm.
Figure 7. Two Sequences from the attB Region Protected by Int from Nuclease Digestion.
The top sequence includes the 15 base common core sequence (bold type). The bottom sequence is from the B′ arm. See the legend to Figure 1 for sequence numbering. (■) Sequences protected from NCS; (□) sequences protected from pancreatic DNAase; (
 ) sequences protected from micrococcal nuclease. All experiments were carried out with a fragment labeled in the top strand (see Figures 1 and 6). (●), (○) or (
) sequences protected from micrococcal nuclease. All experiments were carried out with a fragment labeled in the top strand (see Figures 1 and 6). (●), (○) or (
 ) Bases whose extent of protection is not known; see legend to Figure 5 and Results. (
) Bases whose extent of protection is not known; see legend to Figure 5 and Results. (
 ) A palindromic sequence.
) A palindromic sequence.
Discussion
When Int protein interacts with fragments of DNA from the attP region, two different DNA sequences are preferentially protected from digestion by either pancreatic DNAase or neocarzinostatin. The two regions are similar in size, 30 bases for the protected sequence that is centered on the core of attP and 35 bases for that located 70 bases to the right in the P′ arm. We do not know whether the two regions of the attachment site that bind to Int interact with one another in an important way. Such interaction should be sterically feasible; for example, in nucleosomes, DNA is wrapped around the histone core proteins such that residues separated by 80 bp lie close to one another (Finch et al., 1977). With the cloning of subfragments of the attachment site region (K. Mizuuchi and M. Mizuuchi, personal communication; P.-L. Hsu and A. Landy, unpublished results) the importance of interactions between various parts of the att region should be determined in the near future.
When the polyanion heparin is present, under our experimental conditions, only the sequence in the P′ arm (and not the sequence around the core) remains protected by Int from nuclease digestion. However, the amount of Int must be increased 5–10 fold over that needed to demonstrate protection in the absence of heparin challenge. This might reflect concentration-dependent changes in the way Int protein interacts with itself and/or with the DNA. For example, it may be that several monomers of Int (acting independently or as an oligomer) are required to make a stable open structure similar to that observed for promoter DNA and RNA polymerase (Chamberlin, 1976). Oligomerization of Int molecules is an especially interesting possibility since electron microscopy has revealed clusters of 8–10 Int molecules bound to the attachment site region (D. Hamilton and R. Yuan, personal communication). On the other hand, the present experiments cannot rule out the possibility that the requirement for higher Int concentration is due simply to the titration of heparin by excess Int (see Experimental Procedures).
Regardless of the mechanism responsible for heparin resistance, the different response to challenge by heparin makes it clear that the attP common core and the P′ arm binding sites must contain nonidentical sequence determinants of the interaction with Int. This is in keeping with the fact that the two regions share little sequence homology (see Figure 5 for summary). The occurrence of a single protein binding to two apparently unrelated DNA sequences is unusual. It may be that each sequence binds to a different domain of the Int protein. This is reminiscent of the situation with ribosomal protein S1, which has two nucleic acid binding sites with very different affinities for DNA and RNA (Draper and von Hippel, 1978a, 1978b). Alternatively, a single binding site on Int protein may interact with both the core and P′ arm DNA sequences. In either case, it is not clear that the greater heparin resistance of Int complexes with the P′ arm site reflects a higher association constant. For example, differences in the accessibility of the complexes to heparin could distinguish the two sites (see Pfeffer, Stahl and Chamberlin, 1977).
Although there is no extensive sequence homology between the common core and P′ arm binding sites (see Figure 5), one small homology (TTTTAT) found in the middle of each protected region is interesting because of its similarity to a sequence (TTTAT) conserved in four out of six secondary att site “common core regions” (Landy et al., 1978; Christie and Platt, 1979). This sequence cannot be sufficient for Int binding, however, since it also occurs in a region of the P arm (−61 to −66) that is not protected by Int (see Figure 4). One of the inverted repeats indicated in Figure 5 is of particular interest because of its symmetrical placement within the protected common core region; the bases most strongly protected against NCS digestion include those in the arms of the inverted repeat, whereas less well protected bases lie between them.
It is also interesting to note that the protected site in the P′ arm overlaps with the carboxy terminus of the int gene (defined by two tandem stop codons in the int reading frame at +80 and +83; R. Hoess, K. Bidwell, C. Foeller and A. Landy, manuscript in preparation) (see Figure 5). This region also includes several sequence features characteristic of transcription termination sites (Landy and Ross, 1977). Thus binding of Int at this site might provide an additional means of regulating intracellular levels of Int protein. If such a mechanism exists, it is not likely to be a simple interference with transcription. In λ lysogens where expression of trpB results from transcription initiated within the prophage and extending outward through int, no consistent correlation between levels of Int and trpB enzyme has been observed (Campbell et al., 1977). In addition, expression of gal genes located downstream from int is not affected by structural mutations in the int gene (M. E. Gottesman, personal communication).
Int protection of a DNA fragment from the attB region is quite different from that observed with fragments from the attP region. We have not found any condition under which Int affords protection to attB DNA in the presence of heparin. In the absence of heparin challenge some of the attB core sequence is protected by Int, but this sequence is about half the size of the corresponding region protected in attP-containing fragments. Thus the 15 base core sequence is not the sole determinant of interactions of the core region with Int. From the present experiments we cannot determine whether the difference in Int protection of the attB and attP common core regions is due to sequences immediately adjacent to the core or whether the presence or absence of the P′ arm site is the distinguishing factor. There is little which can be said at this point about the second, less strongly protected region found in the B′ arm (see Figures 6 and 7). However, experiments with cloned and shortened att sites (P.-L. Hsu and A. Landy, unpublished observations) should soon reveal whether it has a role in Int-dependent recombination or whether it is merely one of a number of sequences which may possess varying degrees of affinity for the Int protein.
The protection experiments reported in this paper are carried out in the absence of the additional factors required for integrative recombination: spermidine, host protein and a second attachment site. In addition, we have assumed that the protection of DNA afforded by our preparation of highly active, nearly homogeneous Int protein reflects the properties of active Int molecules rather than denatured Int or contaminant proteins. Moreover, we have no assurance that the fragments whose protection by Int we have studied encompass all the sequences required for full attachment site function. Despite these limitations, we believe that the following are reasonable interpretations of our data. First, the P′ arm is believed to have a singular role in λ site-specific recombination. For example, in vegetative crosses involving different combinations of attachment sites, efficient recombination often occurs when one of the attachment sites carries the P′ arm but is reduced or does not occur when the P′ arm is absent (in such cases, Int is supplied from a helper phage) (Parkinson, 1971). The results reported here establish that Int interacts in a unique manner with the P′ arm, forming a heparin-insensitive complex with the sequence at position 50–85—a short but significant distance from the site of recombination crossover. Second, the protection of a 30 base segment which includes the common core region demonstrates for the first time the interaction of Int with the bases of the crossover locus of attP. This result taken together with the finding that Int is a topoisomerase (that is, has the ability to break and reseal strands of DNA) (Kikuchi and Nash, 1979) suggests that Int itself catalyzes the strand exchange. Finally, the finding that Int protects a specific region of attB at the same concentration and with the same efficiency as it protects attP suggests that Int may recognize both sites involved in integrative recombination. Further studies on the Int protection of attB in the presence of attP, or portions of the attP region, should help to clarify the interaction of Int with recombining att sites.
Experimental Procedures
Preparation of Labeled Restriction Fragments
Two restriction fragments derived from the attP region were used in these experiments, one labeled at the 5′ end of the bottom strand, the other labeled at the 5′ end of the top strand. The first, a Hinf I (−114)-Mnl I (+115) fragment (see Figure 1 and Landy and Ross, 1977), was cloned between the Hind III and Bam HI sites on the plasmid pBR322. A Hind III linker (Collaborative Research) was put onto the Hinf I site and a Bam HI linker onto the Mnl I site (plasmid pPH3; P.-L. Hsu and A. Landy, unpublished results). A double-stranded fragment, labeled in the bottom strand, was prepared by digesting the pPH3 plasmid at the Bam HI site and labeling with polynucleotide kinase (Boehringer-Mannheim) and γ-32P-ATP (New England Nuclear), using modifications of the method described by Maxam and Gilbert (1977) as described previously (Bidwell and Landy, 1979). The plasmid was then digested with Hind III and the Hind III-Bam HI fragment was gel-purified. The other fragments, from either the attP or the attB regions (shown in Figure 1), were purified from the appropriate Hind II + III primary fragments from phage λ as described previously (Landy and Ross, 1977). The primary fragment was first digested with the restriction enzyme, generating the end to be 32P-labeled. The digest was labeled as described above and digested with the second restriction enzyme, and the fragment was purified on a 5% acrylamide gel. After gel elution, DNA fragments were separated from soluble gel material by passage through BND cellulose columns (Serva). The restriction nucleases used to prepare the labeled fragments were obtained from New England Biolabs.
DNAase Footprinting Experiments
The DNA-Int binding reactions were carried out based on the conditions previously described (Kikuchi and Nash, 1978a). Approximately 0.01 pmole (1–2 ng) of labeled DNA restriction fragment (containing approximately 0.1 μg of carrier RNA; Type VI, Sigma) was incubated with varying amounts of purified Int protein (1.25–10 μg/ml) in 20–40 μl of binding buffer [50 mM Tris-Cl (pH 7.4), 60 mM KCl, 2 mg/ml bovine serum albumin, 1 mM MgCl2, 1 mM β-mercaptoethanol, 10% glycerol], Int protein was prepared as described before (Kikuchi and Nash, 1978b); material of >95% homogeneity from two different preparations gave similar results. The molar ratio of Int: att fragment in the binding reactions varied from approximately 50:1 to 500:1. Immediately prior to use, dilutions of Int protein were prepared in binding buffer containing 0.6 M KCl and no MgCl2. The binding mixture was incubated at 20°C for 20 min. Heparin (Sigma) was then added to some samples at a final concentration of 5 μg/ml and incubated for 2 min. Assuming an average molecular weight of 12,000 for heparin, this concentration results in a molar ratio of heparin:Int which varies from approximately 10:1 to 1:1. Pancreatic DNAase (Worthington, DPFF) was then added to a concentration of 10μg/ml and samples were incubated for 10 min at 20°C, followed by immediate extraction with 50 μl of water-saturated phenol in the presence of 3 μg of sonicated salmon sperm carrier DNA and 5 mM EDTA. Sodium acetate was added to a concentration of 0.4 M and the DNA was precipitated with 3 vol of 95% ethanol. The ethanol-washed DNA pellet was resuspended in 6 μl of gel loading buffer (0.05 N NaOH, 0.5 mM EDTA, 7 M urea and 0.5% xylene cyanol and bromophenol blue).
Neocarzinostatin Footprint Experiments
DNA-Int binding reactions were carried out as described above, except that the binding buffer contained 1 mM EDTA, 10 mM β-mercaptoethanol and no MgCl2. Neocarzinostatin (Kayaku Antibiotics, provided by B. Haseltine) was added to a final concentration of 200 μg/ml and digestion was carried out at 20°C for 8–10 min. Digestions were quenched and samples processed as described above.
Micrococcal Nuclease Footprint Experiments
Binding reactions were carried out in binding buffer as described for pancreatic DNAase experiments, but containing 1 mM CaCl2 and no MgCl2. After a 20 min binding reaction, micrococcal nuclease (Worthington) was added to a final concentration of 100 ng/ml and the mixture was incubated at 20°C for 5 min. Reactions were quenched and samples processed as described above.
Restriction Fragment Filter Binding
DNA-Int binding reactions were carried out as described above for neocarzinostatin footprints. Following the 20 min binding reaction (and heparin challenge as indicated) the samples were digested with NCS. The digested samples were diluted with 1 ml of binding buffer without bovine serum albumin and passed through prewashed 13 mm nitrocellulose filters (Schleicher and Schuell) at a rate of ~2 ml/min. Filters were washed and the DNA was eluted from the filters in 0.3 ml 10 mM Tris-Cl (pH 7.9), 1 mM EDTA, 0.5 M NaCl and 0.1% Sarkosyl containing a trace of phenol. Eluted DNA was phenol-extracted and ethanol-precipitated after addition of carrier DNA.
DNA Sequence Markers and Electrophoresis Conditions
Sequence markers were prepared as described by Maxam and Gilbert (1977), using hydrazine to generate fragments specifically cleaved at T and C residues, and dimethylsulfate reaction followed by incubation at 0°C with perchloric acid to generate fragments cleaved at A and G residues. The sequencing gels used were 10% acrylamide-8 M urea, were 0.5 mm in thickness and either 17 inches (standard) or 29 inches in length. The electrophoresis buffer used was 45 mM Tris-borate (pH 8.3), 1.25 mM EDTA. Samples were loaded onto the gels in 0.05 N NaOH, 0.5 mM EDTA, 7 M Urea and 0.05% xylene cyanol and bromophenol blue. Electrophoresis was carried out at 1500–2000 V. Gels were exposed at −70°C to Kodak XR-5 X-ray film, with or without DuPont Lightning Plus Intensifying Screens.
Acknowledgments
We thank Bill Haseltine for the gift of NCS; Jeff Miller, Alan Maxam, Bill Haseltine and Bob Weisberg for helpful discussions; and David Galas, Albert Schmitz, Bob Yuan, Kiyoshi Mizuuchi, Nancy Kleckner and Max Gottesman for communicating unpublished results. We also thank Julie Smith and Eric Johnson for expert technical assistance. This work was supported by a grant from the NIH and a grant from the National Foundation-March of Dimes. A.L. is a Faculty Research Associate of the American Cancer Society.
References
- Bernardi A, Gaillard C, Bernardi G. The specificity of five DNAases as studied by the analysis of 5′-terminal doublets. Eur J Biochem. 1975;52:451–457. doi: 10.1111/j.1432-1033.1975.tb04013.x. [DOI] [PubMed] [Google Scholar]
- Bidwell K, Landy A. Structural features of λ site-specific recombination at a secondary att site in ga/T. Cell. 1979;16:397–406. doi: 10.1016/0092-8674(79)90015-1. [DOI] [PubMed] [Google Scholar]
- Campbell A, Heffernan L, Hu S-L, Szybalski W. The integrase promoter of bacteriophage lambda. In: Bukhari AI, Shapiro JA, Adhya SL, editors. DNA Insertion Elements, Plasmids, and Episomes. New York: Cold Spring Harbor Laboratory; 1977. pp. 375–379. [Google Scholar]
- Chamberlin MJ. RNA polymerase—an overview. In: Losick R, Chamberlin M, editors. RNA Polymerase. New York: Cold Spring Harbor Laboratory; 1976. pp. 35–37. [Google Scholar]
- Christie GE, Platt T. A secondary attachment site for bacteriophage λ in trpC of E. coli. Cell. 1979;16:407–413. doi: 10.1016/0092-8674(79)90016-3. [DOI] [PubMed] [Google Scholar]
- D’Andrea AD, Haseltine W. Sequence specific cleavage of DNA by the anti-tumor antibiotics neocarzinostatin and bleomycin. Proc Nat Acad Sci USA. 1978;75:3608–3612. doi: 10.1073/pnas.75.8.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper DE, von Hippel PH. Nucleic acid binding properties of Escherichia coli ribosomal protein S1.I Structure and interactions of binding site I. J Mol Biol. 1978a;122:321–338. doi: 10.1016/0022-2836(78)90193-6. [DOI] [PubMed] [Google Scholar]
- Draper DE, von Hippel PH. Nucleic acid binding properties of Escherichia coli ribosomal protein S1. II Co-operativity and specificity of binding site II. J Mol Biol. 1978b;122:339–359. doi: 10.1016/0022-2836(78)90194-8. [DOI] [PubMed] [Google Scholar]
- Finch JT, Lutter LC, Rhodes D, Brown RS, Rushton B, Levitt M, Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977;269:29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- Galas DJ, Schmitz A. DNAase footprinting: a simple method for the detection of protein-DNA binding specificity. Nucl Acids Res. 1978;5:3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W, Maxam A, Mirzabekov A. Contacts between the lac repressor and DNA revealed by methylation. In: Kjelgaard NO, Maaløe O, editors. In Control of Ribosome Synthesis, The Alfred Benzon Symposium. IX. Copenhagen; Munksgaard: 1976. pp. 139–148. [Google Scholar]
- Gottesman ME, Weisberg RA. Prophage insertion and excision. In: Hershey AD, editor. The Bacteriophage Lambda. New York: Cold Spring Harbor Laboratory; 1971. pp. 113–138. [Google Scholar]
- Hatayama T, Goldberg I, Takeshita M, Grollman A. Nucleotide specificity in DNA scission by neocarzinostatin. Proc Nat Acad Sci USA. 1978;75:3603–3607. doi: 10.1073/pnas.75.8.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y, Nash H. Purification and properties of a protein involved in genetic recombination: the bacteriophage λ int gene product. J Biol Chem. 1978a;253:7149–7157. [PubMed] [Google Scholar]
- Kikuchi Y, Nash H. Integrative recombination of phage lambda: requirement for supertwisted DNA in vivo and characterization of Int. Cold Spring Harbor Symp Quant Biol. 1978b;43:1099–1109. doi: 10.1101/sqb.1979.043.01.122. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Nash H. Nicking-closing activity associated with bacteriophage λ int gene product. Proc Nat Acad Sci USA. 1979;76:3760–3764. doi: 10.1073/pnas.76.8.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N. DNA sequence analysis of Tn10 insertions: origin and role of 9 bp flanking repetitions during Tn10 translocation. Cell. 1979;16:711–720. doi: 10.1016/0092-8674(79)90087-4. [DOI] [PubMed] [Google Scholar]
- Kotewicz M, Chung S, Takeda Y, Echols H. Characterization of the integration protein of bacteriophage λ as a site-specific DNA-binding protein. Proc Nat Acad Sci USA. 1977;74:1511–1515. doi: 10.1073/pnas.74.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landy A, Ross W. Viral integration and excision: structure of the lambda att sites. Science. 1977;197:1147–1160. doi: 10.1126/science.331474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landy A, Hoess RH, Bidwell K, Ross W. Site-specific recombination in bacteriophage λ—structural features of recombining sites. Cold Spring Harbor Symp Quant Biol. 1978;43:1089–1097. doi: 10.1101/sqb.1979.043.01.121. [DOI] [PubMed] [Google Scholar]
- McConnell DJ, Searcy DG, Sutcliffe JG. A restriction enzyme That from the thermophilic mycoplasma Thermoplasma acidophilum. Nucl Acids Res. 1978;5:1729–1739. doi: 10.1093/nar/5.6.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A, Gilbert W. A new method for sequencing DNA. Proc Nat Acad Sci USA. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash H. Integration and excision of bacteriophage λ. Curr Topics Microbiol Immunol. 1977;78:171–199. doi: 10.1007/978-3-642-66800-5_6. [DOI] [PubMed] [Google Scholar]
- Parkinson S. Deletion mutants of bacteriophage lambda II. Genetic properties of att-defective mutants. J Mol Biol. 1971;56:385–401. doi: 10.1016/0022-2836(71)90472-4. [DOI] [PubMed] [Google Scholar]
- Pfeffer SR, Stahl SJ, Chamberlin MJ. Binding of Escherichia coli RNA polymerase to T7 DNA. Displacement of holoenzyme from promoter complexes by heparin. J Biol Chem. 1977;252:5403–5407. [PubMed] [Google Scholar]
- Sutcliffe JG, Church GM. The cleavage site of the restriction endonuclease Avall. Nucl Acids Res. 1978;5:2313–2319. doi: 10.1093/nar/5.7.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]


