Abstract
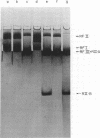
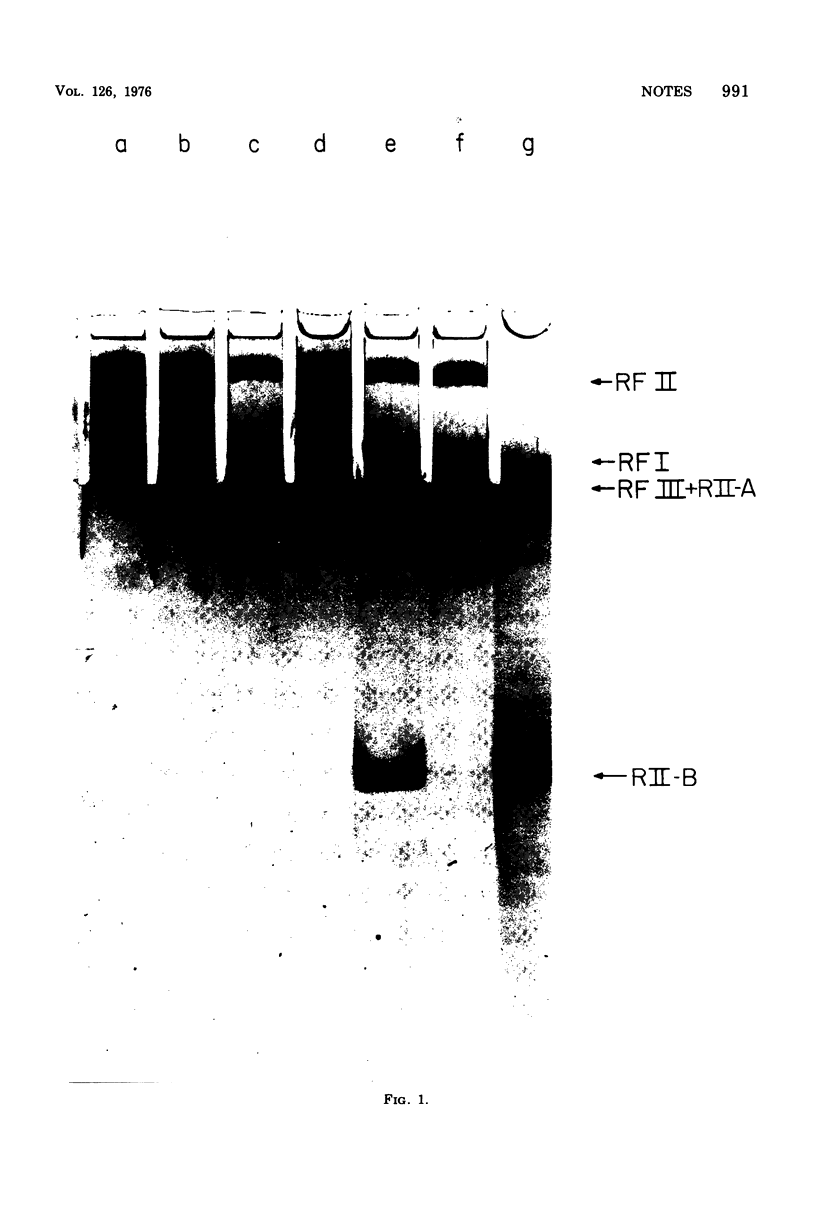
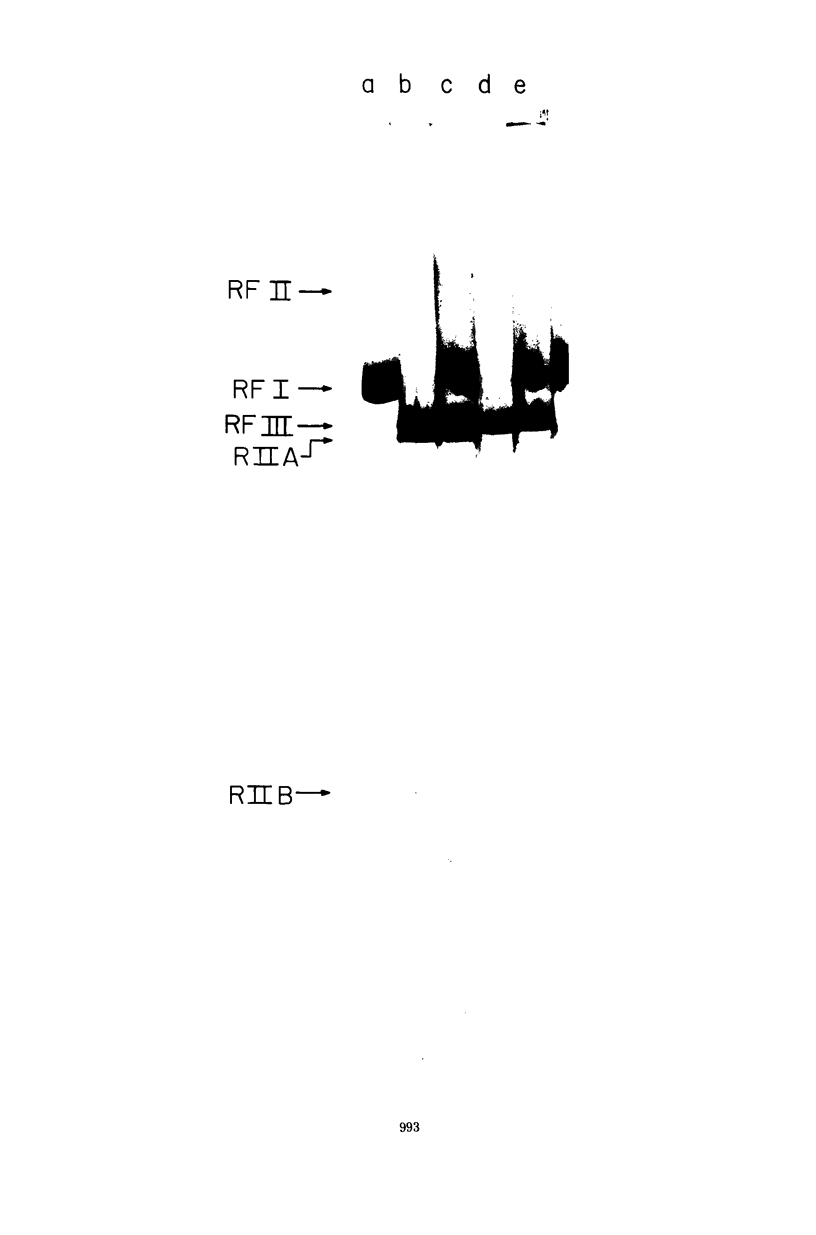
We have analyzed the susceptibility of the deoxyribonucleic acid (DNA) of phage fd replicative form (RF) and of Escherichia coli to in vitro cleavage by purified RII restriction endonuclease (R. Eco RII). The results are summarized as follows: (i) fd, mec- RFI, isolated from infected E. coli K-12 mec- bacteria (a mutant strain lacking DNA-cytosine methylase activity), is cleaved into at least two fragments, whereas fd. mec+ RFI, isolated from the parental mec+ strain, is not cleaved. (ii) E. coli mec- DNA is extensively degraded, whereas mec+ DNA-cytosine methylase acts as an RII modification enzyme.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ARBER W., MORSE M. L. HOST SPECIFICITY OF DNA PRODUCED BY ESCHERICHIA COLI. VI. EFFECTS ON BACTERIAL CONJUGATION. Genetics. 1965 Jan;51:137–148. doi: 10.1093/genetics/51.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskin B., Lautenberger J. A., Linn S. Host-controlled modification and restriction of bacteriophage T7 by escherichia coli B. J Virol. 1973 Jun;11(6):1020–1023. doi: 10.1128/jvi.11.6.1020-1023.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S., Gold E., Plotnik A. Methylation of cytosine residues in DNA controlled by a drug resistance factor (host-induced modification-R factors-N 6 -methyladenine-5-methylcytosine). Proc Natl Acad Sci U S A. 1972 Jan;69(1):187–190. doi: 10.1073/pnas.69.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S. Plasmid-controlled variation in the content of methylated bases in bacteriophage lambda deoxyribonucleic acid. J Virol. 1972 Sep;10(3):356–361. doi: 10.1128/jvi.10.3.356-361.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S. Plasmid-controlled variation in the content of methylated bases in single-stranded DNA bacteriophages M13 and fd. J Mol Biol. 1973 Mar 15;74(4):749–752. doi: 10.1016/0022-2836(73)90064-8. [DOI] [PubMed] [Google Scholar]
- Hattman S., Schlagman S., Cousens L. Isolation of a mutant of Escherichia coli defective in cytosine-specific deoxyribonucleic acid methylase activity and in partial protection of bacteriophage lambda against restriction by cells containing the N-3 drug-resistance factor. J Bacteriol. 1973 Sep;115(3):1103–1107. doi: 10.1128/jb.115.3.1103-1107.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi K., Vovis G. F., Enea V., Zinder N. D. Cleavage map of bacteriophage f1: location of the Escherichia coli B-specific modification sites. J Mol Biol. 1975 Jun 25;95(2):147–165. doi: 10.1016/0022-2836(75)90388-5. [DOI] [PubMed] [Google Scholar]
- Hughes S. G. The sensitivity of bacteriophage lambda DNA to restriction endonuclease RII. J Mol Biol. 1975 Nov 5;98(3):645–647. doi: 10.1016/s0022-2836(75)80093-3. [DOI] [PubMed] [Google Scholar]
- May M. S., Hattaman S. Deoxyribonucleic acid-cytosine methylation by host- and plasmid-controlled enzymes. J Bacteriol. 1975 Apr;122(1):129–138. doi: 10.1128/jb.122.1.129-138.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May M. S., Hattman S. Analysis of bacteriophage deoxyribonucleic acid sequences methylated by host- and R-factor-controlled enzymes. J Bacteriol. 1975 Aug;123(2):768–770. doi: 10.1128/jb.123.2.768-770.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Model P., Horiuchi K., McGill C., Zinder N. D. Template activity of fl RFI cleaved with endonucleases R-Hind, R EcoP1 or R EcoB. Nature. 1975 Jan 10;253(5487):132–134. doi: 10.1038/253132a0. [DOI] [PubMed] [Google Scholar]
- Model P., Zinder N. D. In vitro synthesis of bacteriophage f1 proteins. J Mol Biol. 1974 Feb 25;83(2):231–251. doi: 10.1016/0022-2836(74)90389-1. [DOI] [PubMed] [Google Scholar]
- Schlagman S., Hattman S. Mutants of the N-3 R-factor conditionally defective in hspII modification and deoxyribonucleic acid-cytosine methylase activity. J Bacteriol. 1974 Oct;120(1):234–239. doi: 10.1128/jb.120.1.234-239.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takanami M. Specific cleavage of coliphage fd DNA by five different restriction endonucleases from Haemophilus genus. FEBS Lett. 1973 Aug 15;34(2):318–322. doi: 10.1016/0014-5793(73)80821-x. [DOI] [PubMed] [Google Scholar]
- Vovis G. F., Horiuchi K., Zinder N. D. Endonuclease R-EcoRII restriction of bacteriophage f1 DNA in vitro: ordering of genes V and VII, location of an RNA promotor for gene VIII. J Virol. 1975 Sep;16(3):674–684. doi: 10.1128/jvi.16.3.674-684.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Takano T., Arai T., Nishida H., Sato S. Episome-mediated Transfer of Drug Resistance in Enterobacteriaceae X. Restriction and Modification of Phages by fi R Factors. J Bacteriol. 1966 Aug;92(2):477–486. doi: 10.1128/jb.92.2.477-486.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimori R., Roulland-Dussoix D., Boyer H. W. R factor-controlled restriction and modification of deoxyribonucleic acid: restriction mutants. J Bacteriol. 1972 Dec;112(3):1275–1279. doi: 10.1128/jb.112.3.1275-1279.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]