Abstract
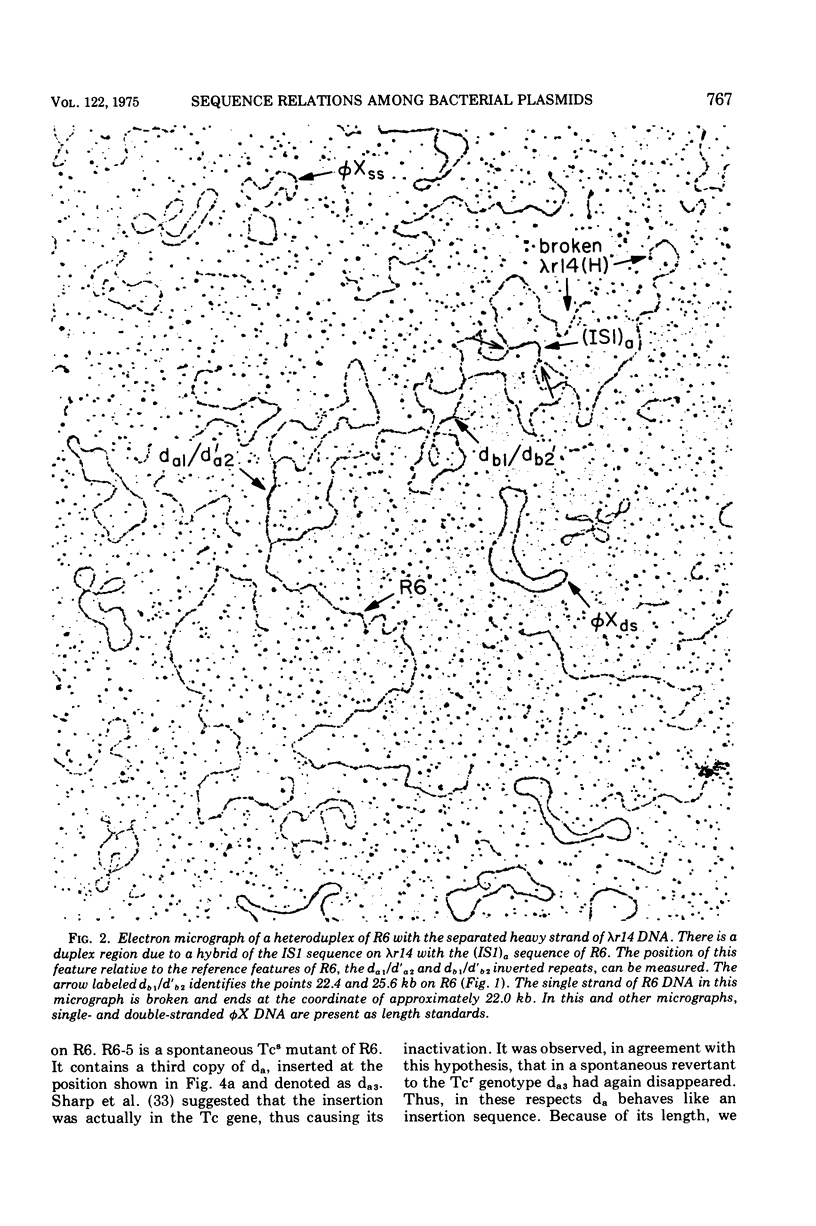
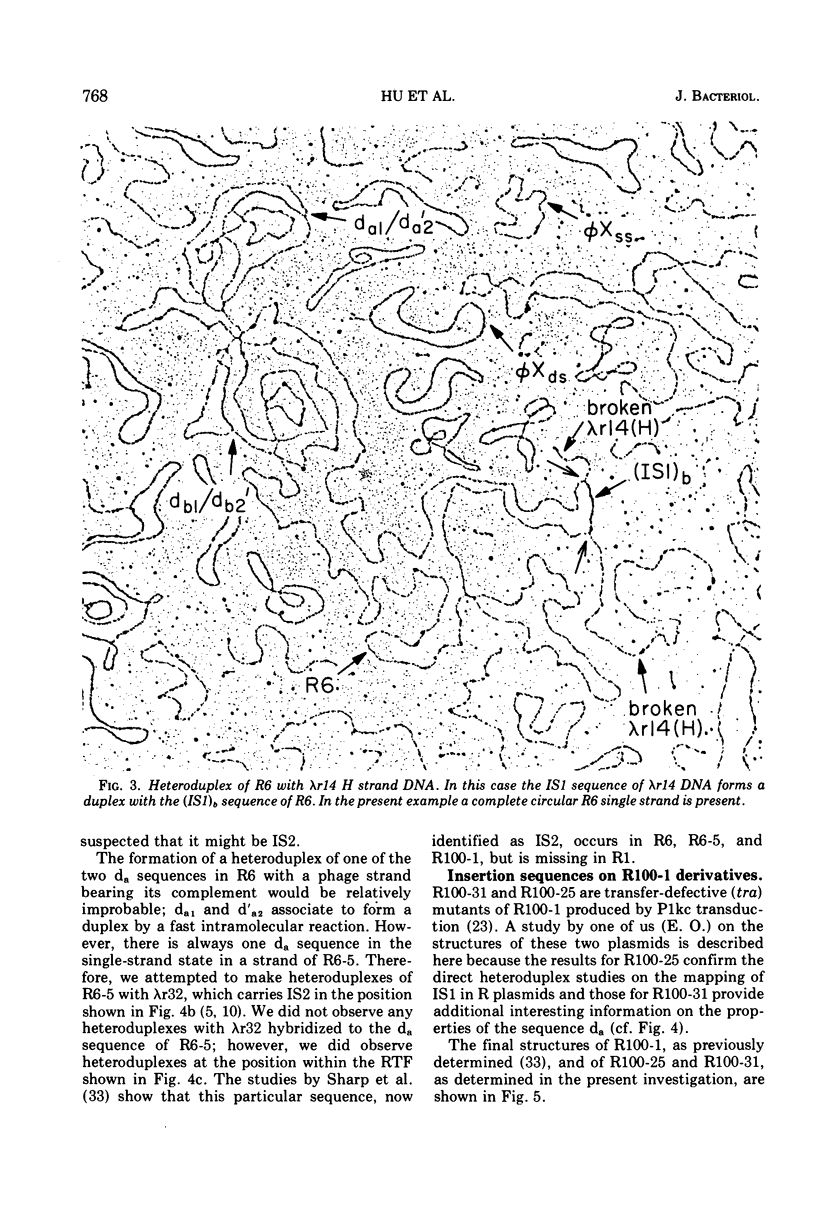
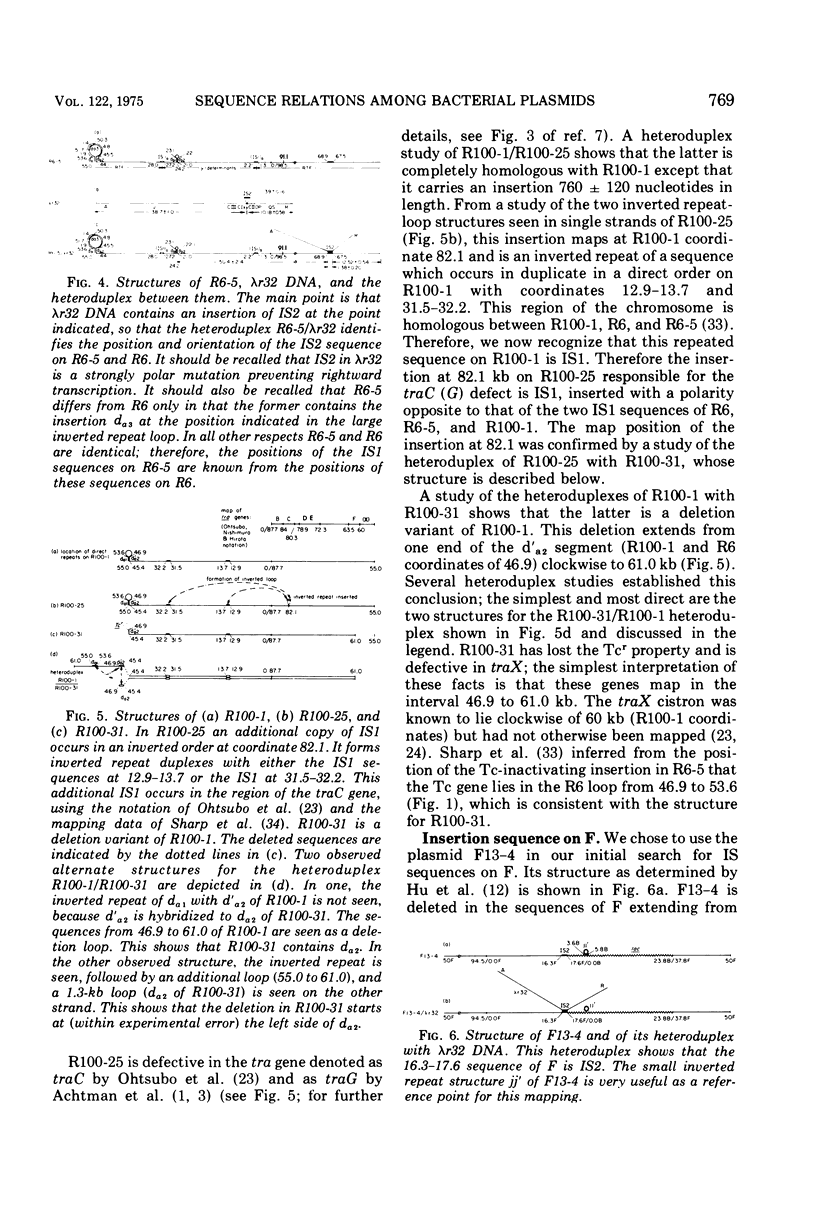
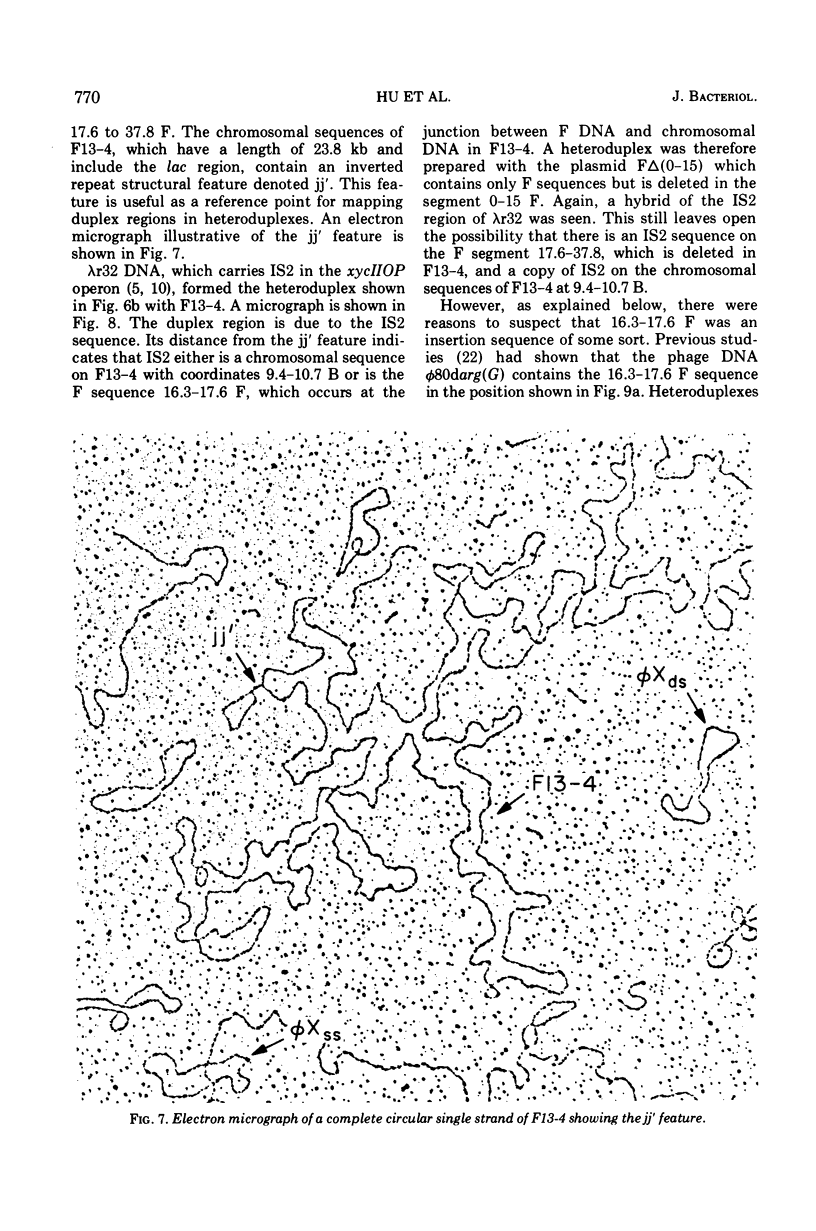
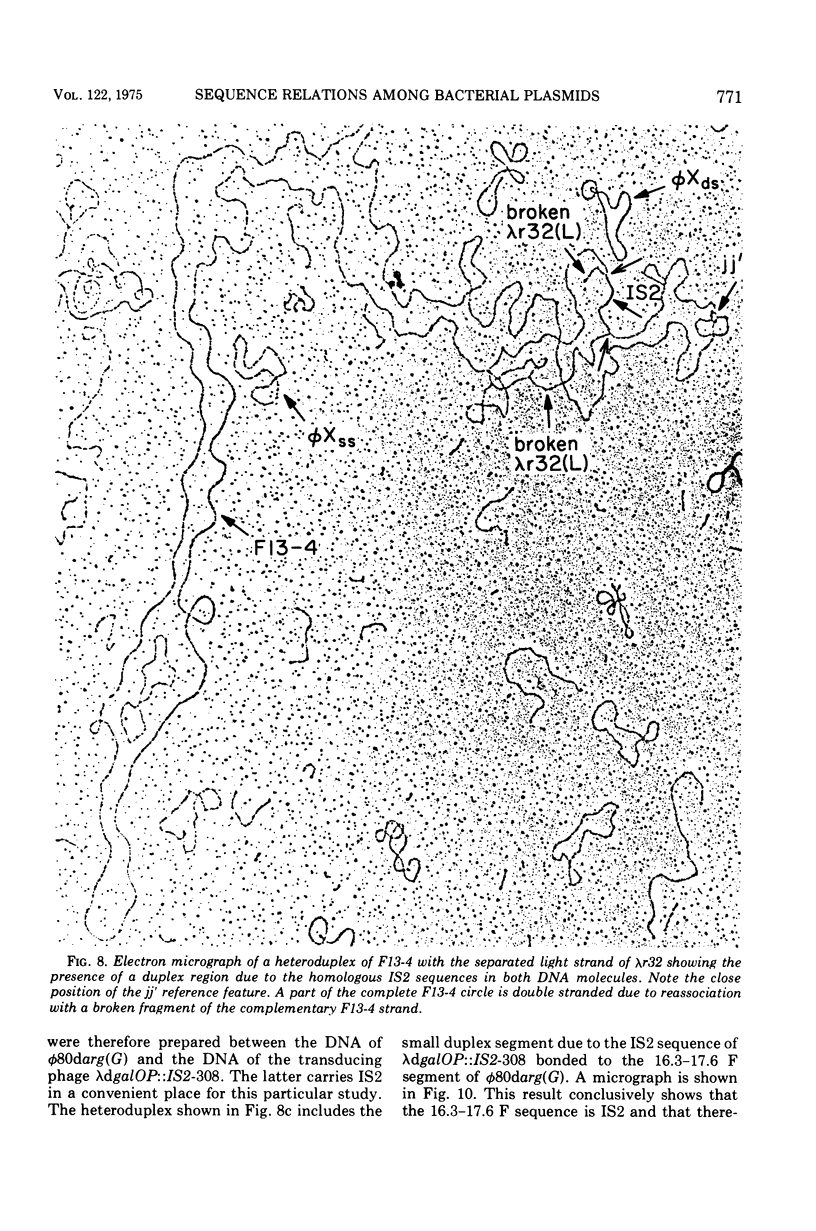
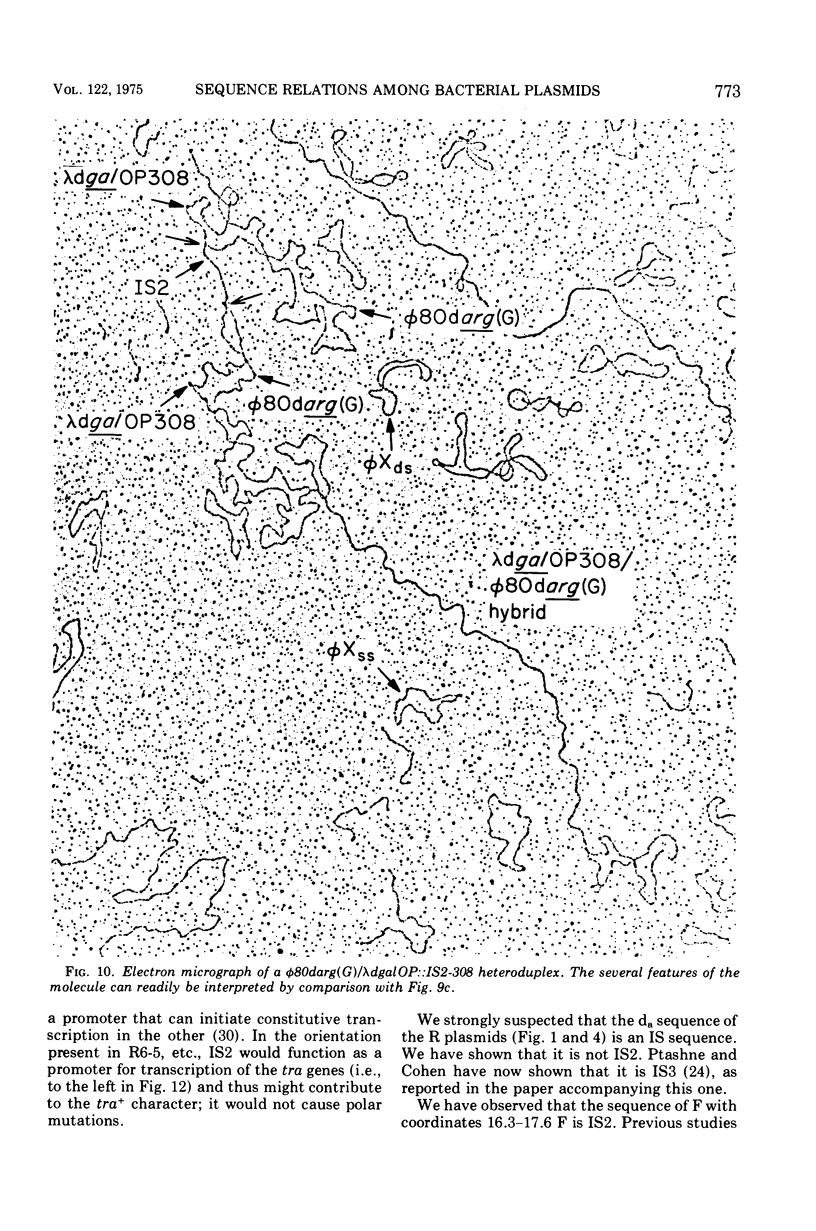
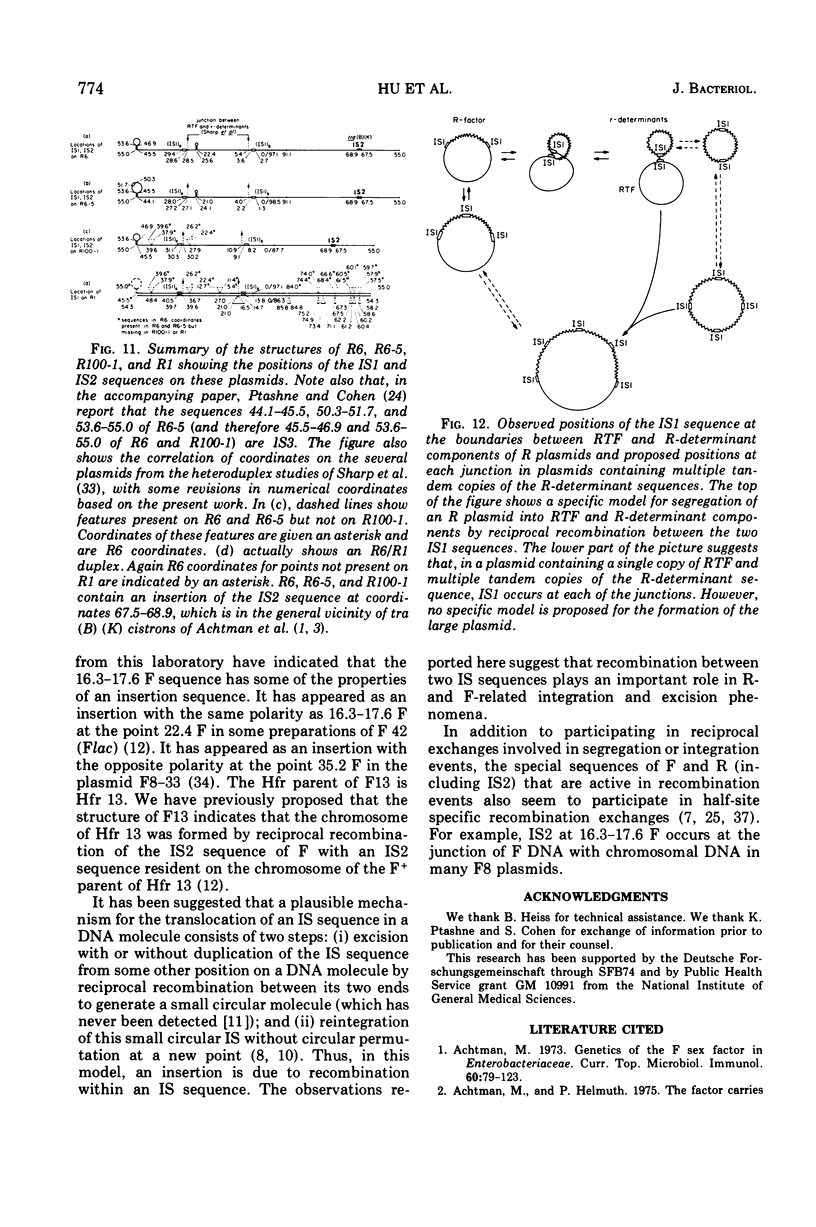
Heteroduplex experiments between the plasmid R6 and one strand of the deoxyribonucleic acid (DNA) of a lambda phage carrying the insertion sequence IS1 show that IS1 occurs on R6 at the two previously mapped junctions of resistance transfer factor (RTF) DNA with R-determinant DNA. From previous heteroduplex experiments, it then follows that IS1 occurs at the same junctions in R6-5, R100-1, and R1 plasmids. Heteroduplex experiments with the DNA from a lambda phage carrying the insertion sequence IS2 show that one copy of IS2 occurs in R6, R6-5, and R100-1 (but not R1) at a point within the RTF with coordinates 67.5 TO 68.9 kilobase units (kb). In an accompanying paper, Ptashne and Cohen (1975) show that the insertion sequence IS3 occurs on R6 and R6-5. R100-25, a traC mutant, differs from its parent R100-1 only in that it contains an additional copy of IS1 inserted within the tra gene region of 82.1 kb. R100-31, atraX, TC-s mutant of R100-1, is deleted in R100-1 sequences starting at one of the IS3 termini (46.9 kb) and extending with RTF to 61.0 kb. Heteroduplex studies of F plasmids with the DNA of a lambda phage bearing insertion sequence IS2 show that the sequence of F with coordinates 16.3-17.6F is IS2. The occurrence of IS1 at the two junctions of R-determinant DNA and RTF DNA in R plasmids provides a structural basis to explain the mechanism of the previously observed formation of molecules containing one RTF unit and several tandem copies of the R-determinant unit, when R plasmids in Proteus mirabilis are grown in the presence of antibiotics, and the segregation of an R plasmid into an RTF unit and an R-determinant unit. In general, correlation of our results with previous studies shows that insertion sequences play a role in a variety of F- and R-related intra- and intermolecular recombination phenomena.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Achtman M. Genetics of the F sex factor in enterobacteriaceae. Curr Top Microbiol Immunol. 1973;60:79–123. doi: 10.1007/978-3-642-65502-9_3. [DOI] [PubMed] [Google Scholar]
- Achtman M., Willetts N., Clark A. J. Beginning a genetic analysis of conjugational transfer determined by the F factor in Escherichia coli by isolation and characterization of transfer-deficient mutants. J Bacteriol. 1971 May;106(2):529–538. doi: 10.1128/jb.106.2.529-538.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhya S. L., Shapiro J. A. The galactose operon of E. coli K-12. I. Structural and pleiotropic mutations of the operon. Genetics. 1969 Jun;62(2):231–247. doi: 10.1093/genetics/62.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachet P., Eisen H., Rambach A. Mutations of coliphage lambda affecting the expression of replicative functions O and P. Mol Gen Genet. 1970;108(3):266–276. doi: 10.1007/BF00283357. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Miller C. A. Non-chromosomal antibiotic resistance in bacteria. II. Molecular nature of R-factors isolated from Proteus mirabilis and Escherichia coli. J Mol Biol. 1970 Jun 28;50(3):671–687. doi: 10.1016/0022-2836(70)90092-6. [DOI] [PubMed] [Google Scholar]
- Fiandt M., Szybalski W., Malamy M. H. Polar mutations in lac, gal and phage lambda consist of a few IS-DNA sequences inserted with either orientation. Mol Gen Genet. 1972;119(3):223–231. doi: 10.1007/BF00333860. [DOI] [PubMed] [Google Scholar]
- Hirsch H. J., Saedler H., Starlinger P. Insertion mutations in the control region of the galactose operon of E. coli. II. Physical characterization of the mutations. Mol Gen Genet. 1972;115(3):266–276. doi: 10.1007/BF00268890. [DOI] [PubMed] [Google Scholar]
- Hirsch H. J., Starlinger P., Brachet P. Two kinds of insertions in bacterial genes. Mol Gen Genet. 1972;119(3):191–206. doi: 10.1007/BF00333858. [DOI] [PubMed] [Google Scholar]
- Hu S., Ohtsubo E., Davidson N. Electron microscopic heteroduplex studies of sequence relations among plasmids of Escherichia coli: structure of F13 and related F-primes. J Bacteriol. 1975 May;122(2):749–763. doi: 10.1128/jb.122.2.749-763.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan E., Saedler H., Starlinger P. O0 and strong-polar mutations in the gal operon are insertions. Mol Gen Genet. 1968;102(4):353–363. doi: 10.1007/BF00433726. [DOI] [PubMed] [Google Scholar]
- Jordan E., Saedler H., Starlinger P. Strong-polar mutations in the transferase gene of the galactose operon in E.coli. Mol Gen Genet. 1967;100(3):296–306. doi: 10.1007/BF00381825. [DOI] [PubMed] [Google Scholar]
- Malamy M. H., Fiandt M., Szybalski W. Electron microscopy of polar insertions in the lac operon of Escherichia coli. Mol Gen Genet. 1972;119(3):207–222. doi: 10.1007/BF00333859. [DOI] [PubMed] [Google Scholar]
- Meynell E., Datta N. Mutant drug resistant factors of high transmissibility. Nature. 1967 May 27;214(5091):885–887. doi: 10.1038/214885a0. [DOI] [PubMed] [Google Scholar]
- Michaelis G., Saedler H., Venkov P., Starlinger P. Two insertions in the galactose operon having different sizes but homologous DNA sequences. Mol Gen Genet. 1969 Aug 15;104(4):371–377. doi: 10.1007/BF00334236. [DOI] [PubMed] [Google Scholar]
- NAKAYA R., NAKAMURA A., MURATA Y. Resistance transfer agents in Shigella. Biochem Biophys Res Commun. 1960 Dec;3:654–659. doi: 10.1016/0006-291x(60)90081-4. [DOI] [PubMed] [Google Scholar]
- Otsubo E., Deonier R. C., Lee H. J., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. IV. The F sequences in F14. J Mol Biol. 1974 Nov 15;89(4):565–584. doi: 10.1016/0022-2836(74)90036-9. [DOI] [PubMed] [Google Scholar]
- Otsubo E., Lee H. J., Deonier R. C., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. VI. Mapping of F14 sequences homologous to phi 80dmetBJF and phi 80dargECBH bacteriophages. J Mol Biol. 1974 Nov 15;89(4):599–618. doi: 10.1016/0022-2836(74)90038-2. [DOI] [PubMed] [Google Scholar]
- Otsubo E., Nishimura Y., Hirota Y. Transfer-defective mutants of sex factors in Escherichia coli. I. Defective mutants and complementation analysis. Genetics. 1970 Feb;64(2):173–188. doi: 10.1093/genetics/64.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne K., Cohen S. N. Occurrence of insertion sequence (IS) regions on plasmid deoxyribonucleic acid as direct and inverted nucleotide sequence duplications. J Bacteriol. 1975 May;122(2):776–781. doi: 10.1128/jb.122.2.776-781.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rownd R., Kasamatsu H., Mickel S. The molecular nature and replication of drug resistance factors of the Enterobacteriaceae. Ann N Y Acad Sci. 1971 Jun 11;182:188–206. doi: 10.1111/j.1749-6632.1971.tb30656.x. [DOI] [PubMed] [Google Scholar]
- Rownd R., Mickel S. Dissociation and reassociation of RTF and r-determinants of the R-factor NR1 in Proteus mirabilis. Nat New Biol. 1971 Nov 10;234(45):40–43. doi: 10.1038/newbio234040a0. [DOI] [PubMed] [Google Scholar]
- Saedler H., Besemer J., Kemper B., Rosenwirth B., Starlinger P. Insertion mutations in the control region of the Gal operon of E. coli. I. Biological characterization of the mutations. Mol Gen Genet. 1972;115(3):258–265. doi: 10.1007/BF00268889. [DOI] [PubMed] [Google Scholar]
- Saedler H., Heiss B. Multiple copies of the insertion-DNA sequences IS1 and IS2 in the chromosome of E. coli K-12. Mol Gen Genet. 1973 May 9;122(3):267–277. doi: 10.1007/BF00278602. [DOI] [PubMed] [Google Scholar]
- Saedler H., Reif H. J., Hu S., Davidson N. IS2, a genetic element for turn-off and turn-on of gene activity in E. coli. Mol Gen Genet. 1974;132(4):265–289. doi: 10.1007/BF00268569. [DOI] [PubMed] [Google Scholar]
- Shapiro J. A. Mutations caused by the insertion of genetic material into the galactose operon of Escherichia coli. J Mol Biol. 1969 Feb 28;40(1):93–105. doi: 10.1016/0022-2836(69)90298-8. [DOI] [PubMed] [Google Scholar]
- Shapiro J., Machattie L., Eron L., Ihler G., Ippen K., Beckwith J. Isolation of pure lac operon DNA. Nature. 1969 Nov 22;224(5221):768–774. doi: 10.1038/224768a0. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Cohen S. N., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. II. Structure of drug resistance (R) factors and F factors. J Mol Biol. 1973 Apr 5;75(2):235–255. doi: 10.1016/0022-2836(73)90018-1. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Hsu M. T., Otsubo E., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. I. Structure of F-prime factors. J Mol Biol. 1972 Nov 14;71(2):471–497. doi: 10.1016/0022-2836(72)90363-4. [DOI] [PubMed] [Google Scholar]
- Silver R. P., Falkow S. Specific labeling and physical characterization of R-factor deoxyribonucleic acid in Escherichia coli. J Bacteriol. 1970 Oct;104(1):331–339. doi: 10.1128/jb.104.1.331-339.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starlinger P., Saedler H. Insertion mutations in microorganisms. Biochimie. 1972;54(2):177–185. doi: 10.1016/s0300-9084(72)80102-0. [DOI] [PubMed] [Google Scholar]
- Sugino Y. Mutants of Escherichia coli sensitive to methylene blue and acridines. Genet Res. 1966 Feb;7(1):1–11. doi: 10.1017/s0016672300009423. [DOI] [PubMed] [Google Scholar]
- Tye B. K., Chan R. K., Botstein D. Packaging of an oversize transducing genome by Salmonella phage P22. J Mol Biol. 1974 Jan 5;85(4):485–500. doi: 10.1016/0022-2836(74)90311-8. [DOI] [PubMed] [Google Scholar]







