Abstract
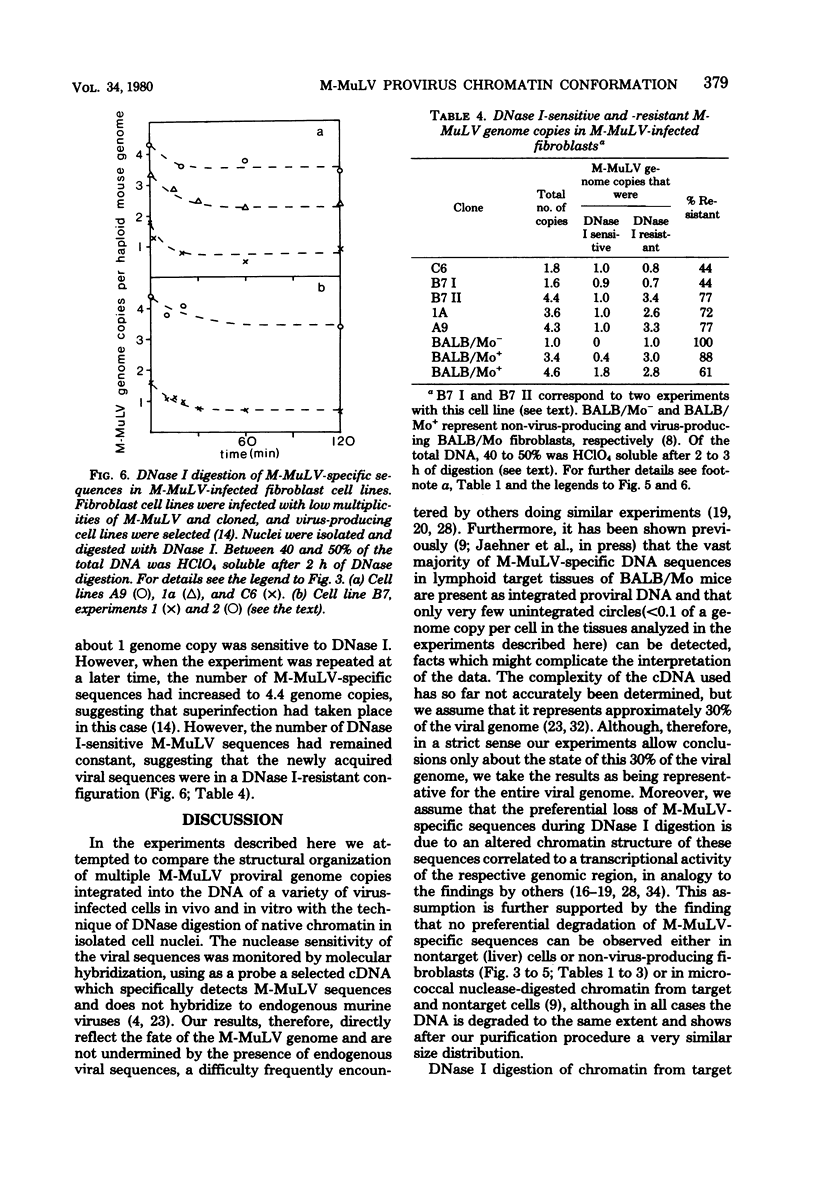
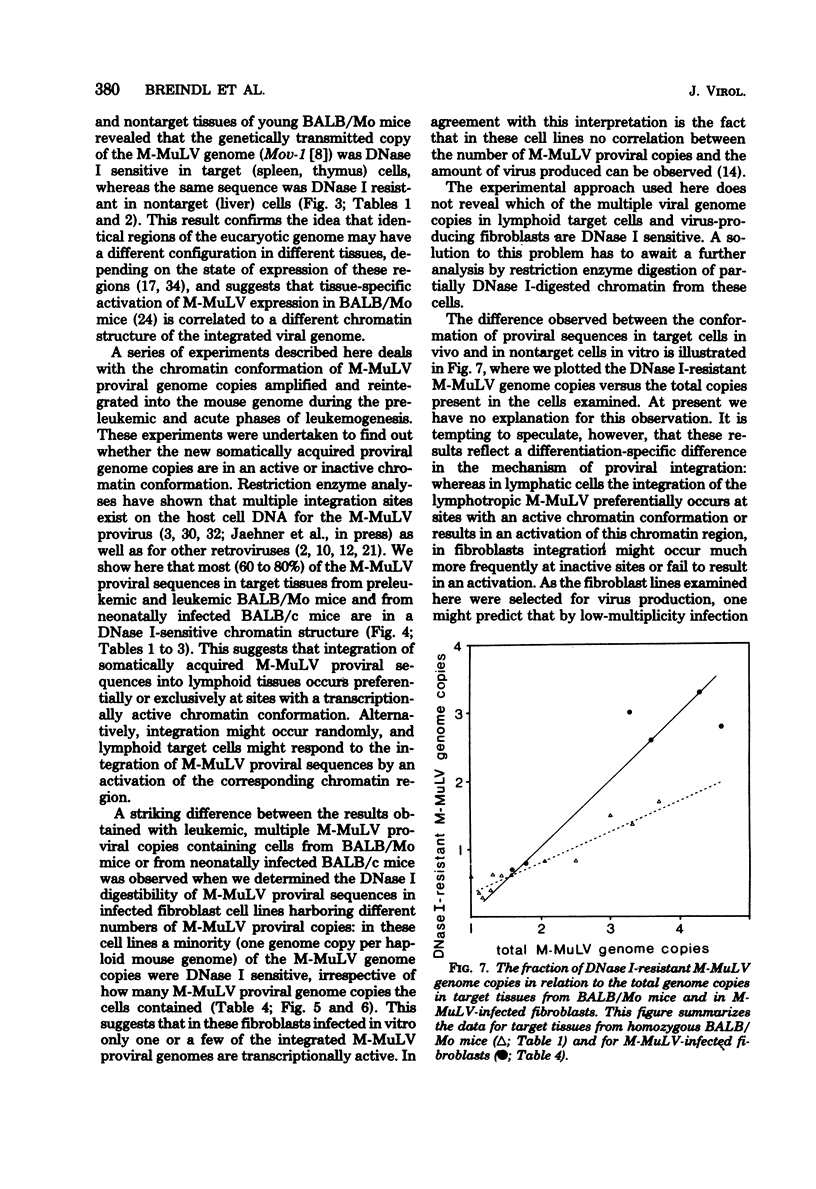
The technique of preferential DNase I digestion of transcriptionally active chromatin regions was used to study the structural organization of integrated Moloney murine leukemia virus (M-MuLV) proviral sequences in various cells carrying integrated viral genomes. BALB/Mo mice, which carry M-MuLV as an endogenous virus at a single Mendelian locus, were used to examine the genetically transmitted viral genome copy and additional M-MuLV sequences acquired somatically during leukemogenesis. It has been shown previously that M-MuLV genome expression in these mice is restricted to lymphatic target tissues. In young homozygous BALB/Mo mice carrying one M-MuLV genome copy per haploid mouse genome in all cells we found that the genetically transmitted viral genome copy was in a preferentially DNase I-sensitive conformation in lymphatic target tissues, whereas in nontarget tissues the same sequence was not preferentially DNase I sensitive. This suggests that the chromatin conformation and the transcriptional activity of the integrated proviral genome are related to and probably determined by the state of cellular differentiation. In target tissues from BALB/Mo mice examined at different ages and in different stages of leukemogenesis the majority of the new somatically acquired M-MuLV sequences were preferentially DNase I digestible. A very similar pattern of DNase I digestibility was observed in target tissues from BALB/c mice exogenously infected with M-MuLV. This shows that in these tissues somatically acquired proviral sequences integrate preferentially or exclusively at sites of the host genome in which they are in a transcriptionally active chromatin conformation. Alternatively, the chromatin structure of the respective host genome region may be changed after the integration of viral DNA. In nontarget tissues from BALB/Mo mice the M-MuLV-specific sequences remained DNase I resistant throughout the lives of the animals. A different pattern of DNase I digestibility was observed in virus-infected cell lines which had been produced by low-multiplicity infection, cloned, and selected for virus production. When cell lines harboring different numbers of M-MuLV proviral copies were examined, it was found that a minority of the proviral sequences (on the average only one M-MuLV genome copy per haploid mouse genome) were preferentially digestible by DNase I, independent of the total number of proviral genome copies present. This suggests that the chromatin conformation of newly acquired proviral sequences is influenced by the state of differentiation of the infected cell or the way infected cells are selected or both.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Astrin S. M. Endogenous viral genes of the White Leghorn chicken: common site of residence and sites associated with specific phenotypes of viral gene expression. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5941–5945. doi: 10.1073/pnas.75.12.5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacheler L. T., Fan H. Multiple integration sites for Moloney murine leukemia virus in productively infected mouse fibroblasts. J Virol. 1979 Jun;30(3):657–667. doi: 10.1128/jvi.30.3.657-667.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns A., Jaenisch R. Increase of AKR-specific sequences in tumor tissues of leukemic AKR mice. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2448–2452. doi: 10.1073/pnas.73.7.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop J. M. Retroviruses. Annu Rev Biochem. 1978;47:35–88. doi: 10.1146/annurev.bi.47.070178.000343. [DOI] [PubMed] [Google Scholar]
- Bloom K. S., Anderson J. N. Fractionation of hen oviduct chromatin into transcriptionally active and inactive regions after selective micrococcal nuclease digestion. Cell. 1978 Sep;15(1):141–150. doi: 10.1016/0092-8674(78)90090-9. [DOI] [PubMed] [Google Scholar]
- Boettiger D. Virogenic nontransformed cells isolated following infection of normal rat kidney cells with B77 strain Rous sarcoma virus. Cell. 1974 Sep;3(1):71–76. doi: 10.1016/0092-8674(74)90042-7. [DOI] [PubMed] [Google Scholar]
- Breindl M., Doehmer J., Willecke K., Dausman J., Jaenisch R. Germ line integration of Moloney leukemia virus: identification of the chromosomal integration site. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1938–1942. doi: 10.1073/pnas.76.4.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breindl M., Jaenisch R. Conformation of Moloney murine leukaemia proviral sequences in chromatin from leukaemic and nonleukaemic cells. Nature. 1979 Jan 25;277(5694):320–322. doi: 10.1038/277320a0. [DOI] [PubMed] [Google Scholar]
- Canaani E., Aaronson S. A. Restriction enzyme analysis of mouse cellular type C viral DNA: emergence of new viral sequences in spontaneous AKR/J lymphomas. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1677–1681. doi: 10.1073/pnas.76.4.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R. J., Felsenfeld G. Structure of chromatin. Nat New Biol. 1971 Jan 27;229(4):101–106. doi: 10.1038/newbio229101a0. [DOI] [PubMed] [Google Scholar]
- Cohen J. C., Varmus H. E. Endogenous mammary tumour virus DNA varies among wild mice and segregates during inbreeding. Nature. 1979 Mar 29;278(5703):418–423. doi: 10.1038/278418a0. [DOI] [PubMed] [Google Scholar]
- Fan H., Baltimore D. RNA metabolism of murine leukemia virus: detection of virus-specific RNA sequences in infected and uninfected cells and identification of virus-specific messenger RNA. J Mol Biol. 1973 Oct 15;80(1):93–117. doi: 10.1016/0022-2836(73)90235-0. [DOI] [PubMed] [Google Scholar]
- Fan H., Jaenisch R., MacIsaac P. Low-multiplicity infection of Moloney murine leukemia virus in mouse cells: effect on number of viral DNA copies and virus production in producer cells. J Virol. 1978 Dec;28(3):802–809. doi: 10.1128/jvi.28.3.802-809.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin. Nature. 1978 Jan 12;271(5641):115–122. doi: 10.1038/271115a0. [DOI] [PubMed] [Google Scholar]
- Flint S. J., Weintraub H. M. An altered subunit configuration associated with the actively transcribed DNA of integrated adenovirus genes. Cell. 1977 Nov;12(3):783–794. doi: 10.1016/0092-8674(77)90277-x. [DOI] [PubMed] [Google Scholar]
- Garel A., Axel R. Selective digestion of transcriptionally active ovalbumin genes from oviduct nuclei. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3966–3970. doi: 10.1073/pnas.73.11.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garel A., Zolan M., Axel R. Genes transcribed at diverse rates have a similar conformation in chromatin. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4867–4871. doi: 10.1073/pnas.74.11.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groudine M., Das S., Neiman P., Weintraub H. Regulation of expression and chromosomal subunit conformation of avian retrovirus genomes. Cell. 1978 Aug;14(4):865–878. doi: 10.1016/0092-8674(78)90342-2. [DOI] [PubMed] [Google Scholar]
- Howk R. S., Anisowicz A., Silverman A. Y., Parks W. P., Scoinick E. M. Distribution of murine type B and type C viral nucleic acid sequences in template active and template inactive chromatin. Cell. 1975 Apr;4(4):321–327. doi: 10.1016/0092-8674(75)90152-x. [DOI] [PubMed] [Google Scholar]
- Hughes S. H., Shank P. R., Spector D. H., Kung H. J., Bishop J. M., Varmus H. E., Vogt P. K., Breitman M. L. Proviruses of avian sarcoma virus are terminally redundant, co-extensive with unintegrated linear DNA and integrated at many sites. Cell. 1978 Dec;15(4):1397–1410. doi: 10.1016/0092-8674(78)90064-8. [DOI] [PubMed] [Google Scholar]
- Jaenisch R. Germ line integration and Mendelian transmission of the exogenous Moloney leukemia virus. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1260–1264. doi: 10.1073/pnas.73.4.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R. Germ line integration of moloney leukemia virus: effect of homozygosity at the m-mulV locus. Cell. 1977 Nov;12(3):691–696. doi: 10.1016/0092-8674(77)90269-0. [DOI] [PubMed] [Google Scholar]
- Jaenisch R. Moloney leukemia virus gene expression and gene amplification in preleukemic and leukemic BALB/Mo mice. Virology. 1979 Feb;93(1):80–90. doi: 10.1016/0042-6822(79)90277-0. [DOI] [PubMed] [Google Scholar]
- Lerner R. A., Wilson C. B., Villano B. C., McConahey P. J., Dixon F. J. Endogenous oncornaviral gene expression in adult and fetal mice: quantitative, histologic, and physiologic studies of the major viral glycorprotein, gp70. J Exp Med. 1976 Jan 1;143(1):151–166. doi: 10.1084/jem.143.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Wilson B., Dixon G. H. Limited action of micrococcal nuclease on trout testis nuclei generates two mononucleosome subsets enriched in transcribed DNA sequences. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1682–1686. doi: 10.1073/pnas.76.4.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panet A., Cedar H. Selective degradation of integrated murine leukemia proviral DNA by deoxyribonucleases. Cell. 1977 Aug;11(4):933–940. doi: 10.1016/0092-8674(77)90304-x. [DOI] [PubMed] [Google Scholar]
- Smith H. S., Gelb L. D., Martin M. A. Detection and quantitation of simian virus 40 genetic material in abortively transformed BALB-3T3 clones (mice-diploid cells-virus equivalents). Proc Natl Acad Sci U S A. 1972 Jan;69(1):152–156. doi: 10.1073/pnas.69.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves R., Lilly F. Interactions between host and viral genomes in mouse leukemia. Annu Rev Genet. 1977;11:277–296. doi: 10.1146/annurev.ge.11.120177.001425. [DOI] [PubMed] [Google Scholar]
- Steffen D., Weinberg R. A. The integrated genome of murine leukemia virus. Cell. 1978 Nov;15(3):1003–1010. doi: 10.1016/0092-8674(78)90284-2. [DOI] [PubMed] [Google Scholar]
- Voordouw G., Eisenberg H. Binding of additional histones to chromatin core particles. Nature. 1978 Jun 8;273(5662):446–448. doi: 10.1038/273446a0. [DOI] [PubMed] [Google Scholar]
- Wu C., Bingham P. M., Livak K. J., Holmgren R., Elgin S. C. The chromatin structure of specific genes: I. Evidence for higher order domains of defined DNA sequence. Cell. 1979 Apr;16(4):797–806. doi: 10.1016/0092-8674(79)90095-3. [DOI] [PubMed] [Google Scholar]
- Wu C., Wong Y. C., Elgin S. C. The chromatin structure of specific genes: II. Disruption of chromatin structure during gene activity. Cell. 1979 Apr;16(4):807–814. doi: 10.1016/0092-8674(79)90096-5. [DOI] [PubMed] [Google Scholar]
- van der Putten H., Terwindt E., Berns A., Jaenisch R. The integration sites of endogenous and exogenous Moloney murine leukemia virus. Cell. 1979 Sep;18(1):109–116. doi: 10.1016/0092-8674(79)90359-3. [DOI] [PubMed] [Google Scholar]


