Abstract
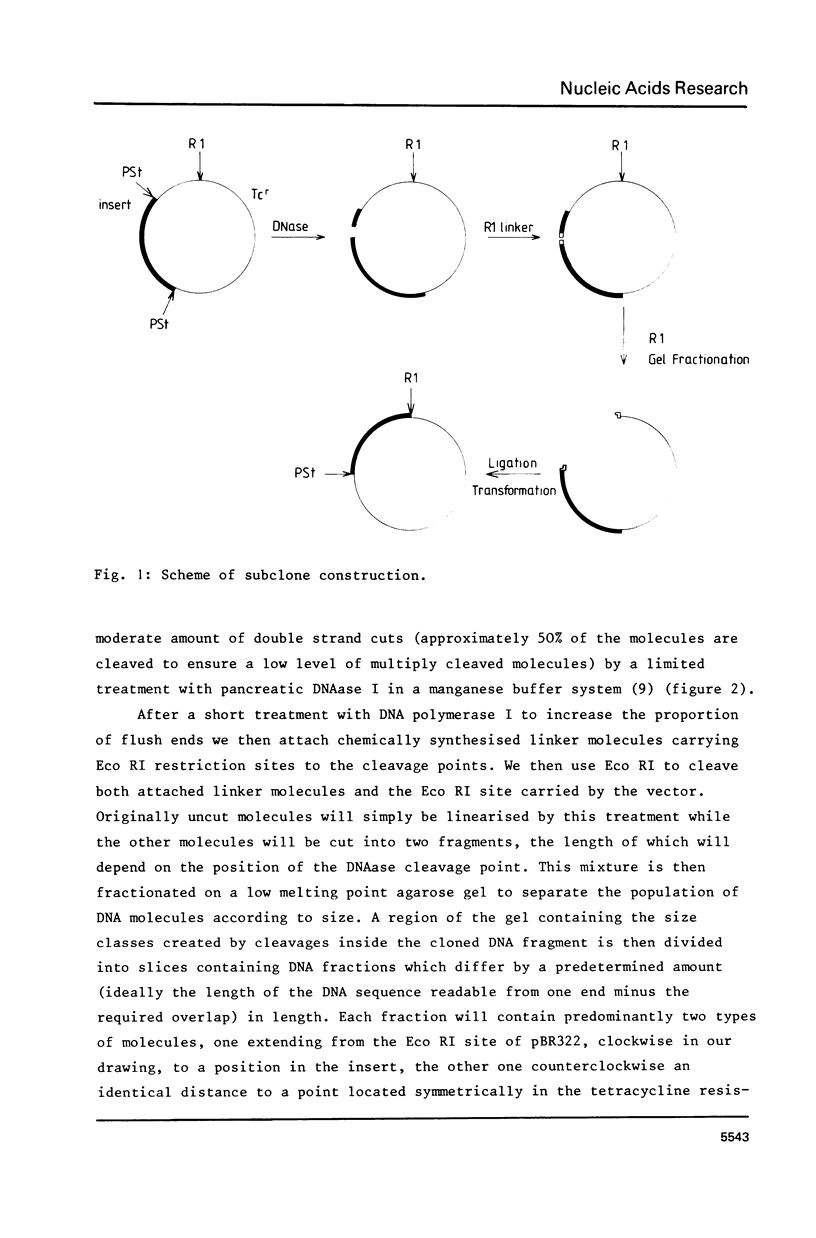
We describe here a new strategy of fragment preparation for sequencing procedures using endlabelled DNA fragments as substrates (2,3) which is directly applicable to DNA fragments cloned into the Pst I site of pBR322, or in modified form, to inserts into the BamH I or Sal I site of the same plasmid. Ordered sets of subclones of predetermined overlap are are generated. These can be sequenced directly without further strand- or fragment separation steps.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Gingeras T. R., Milazzo J. P., Sciaky D., Roberts R. J. Computer programs for the assembly of DNA sequences. Nucleic Acids Res. 1979 Sep 25;7(2):529–545. doi: 10.1093/nar/7.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron F., So M., McCarthy B. J. In vitro mutagenesis of a circular DNA molecule by using synthetic restriction sites. Proc Natl Acad Sci U S A. 1978 Dec;75(12):6012–6016. doi: 10.1073/pnas.75.12.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maat J., Smith A. J. A method for sequencing restriction fragments with dideoxynucleoside triphosphates. Nucleic Acids Res. 1978 Dec;5(12):4537–4545. doi: 10.1093/nar/5.12.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panet A., van de Sande J. H., Loewen P. C., Khorana H. G., Raae A. J., Lillehaug J. R., Kleppe K. Physical characterization and simultaneous purification of bacteriophage T4 induced polynucleotide kinase, polynucleotide ligase, and deoxyribonucleic acid polymerase. Biochemistry. 1973 Dec 4;12(25):5045–5050. doi: 10.1021/bi00749a003. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreier P. H., Cortese R. A fast and simple method for sequencing DNA cloned in the single-stranded bacteriophage M13. J Mol Biol. 1979 Mar 25;129(1):169–172. doi: 10.1016/0022-2836(79)90068-8. [DOI] [PubMed] [Google Scholar]
- Staden R. A strategy of DNA sequencing employing computer programs. Nucleic Acids Res. 1979 Jun 11;6(7):2601–2610. doi: 10.1093/nar/6.7.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]