Abstract
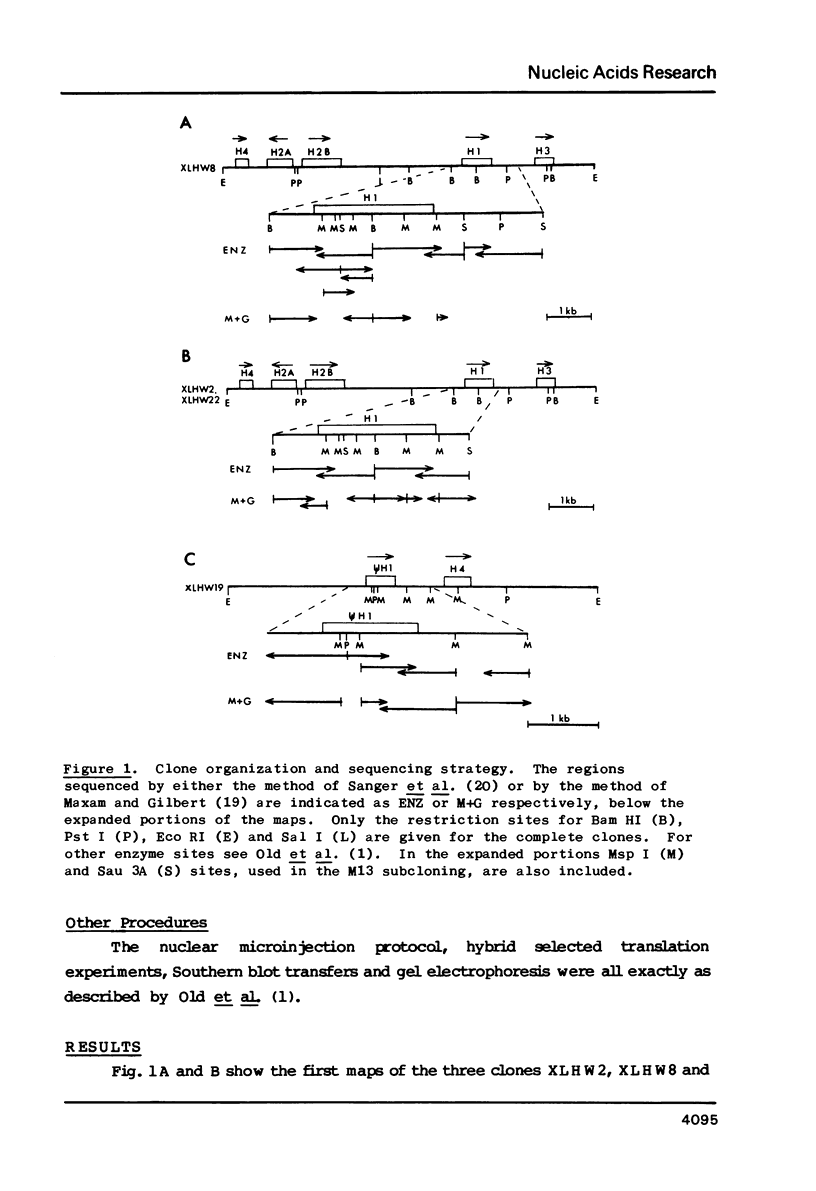
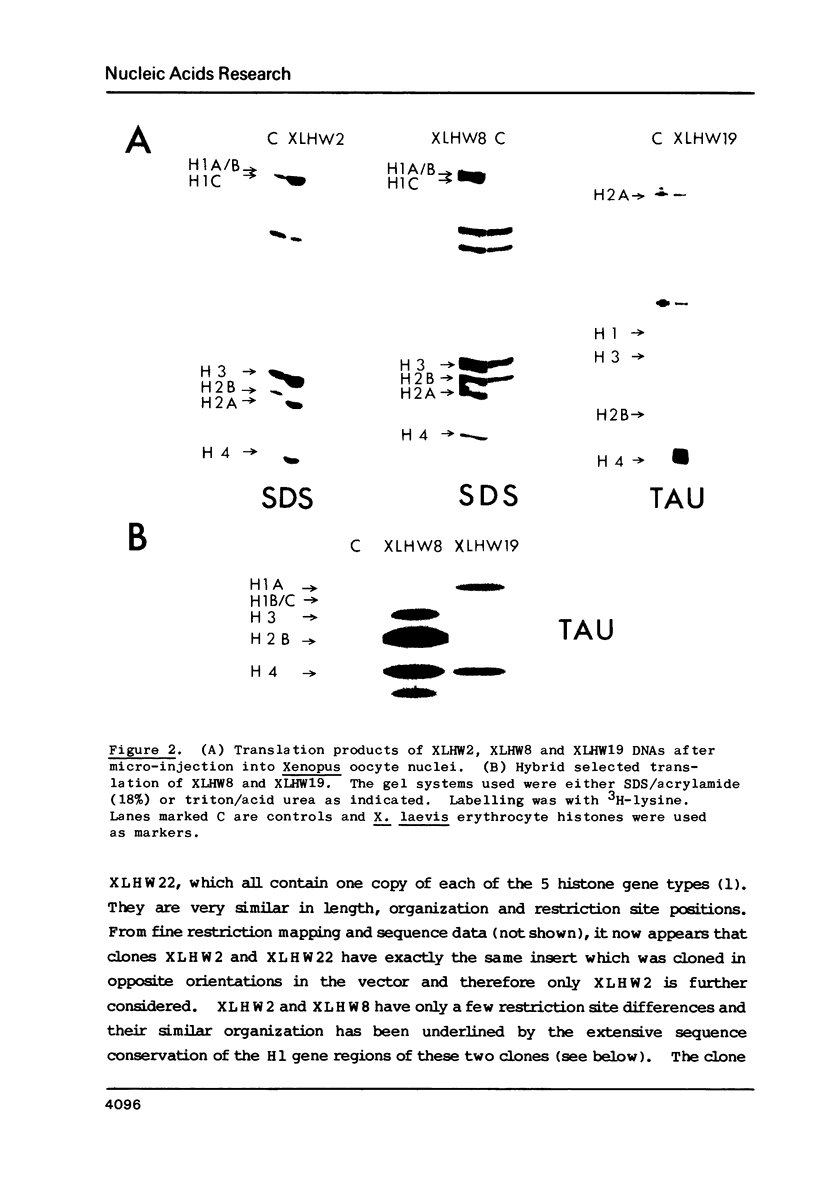
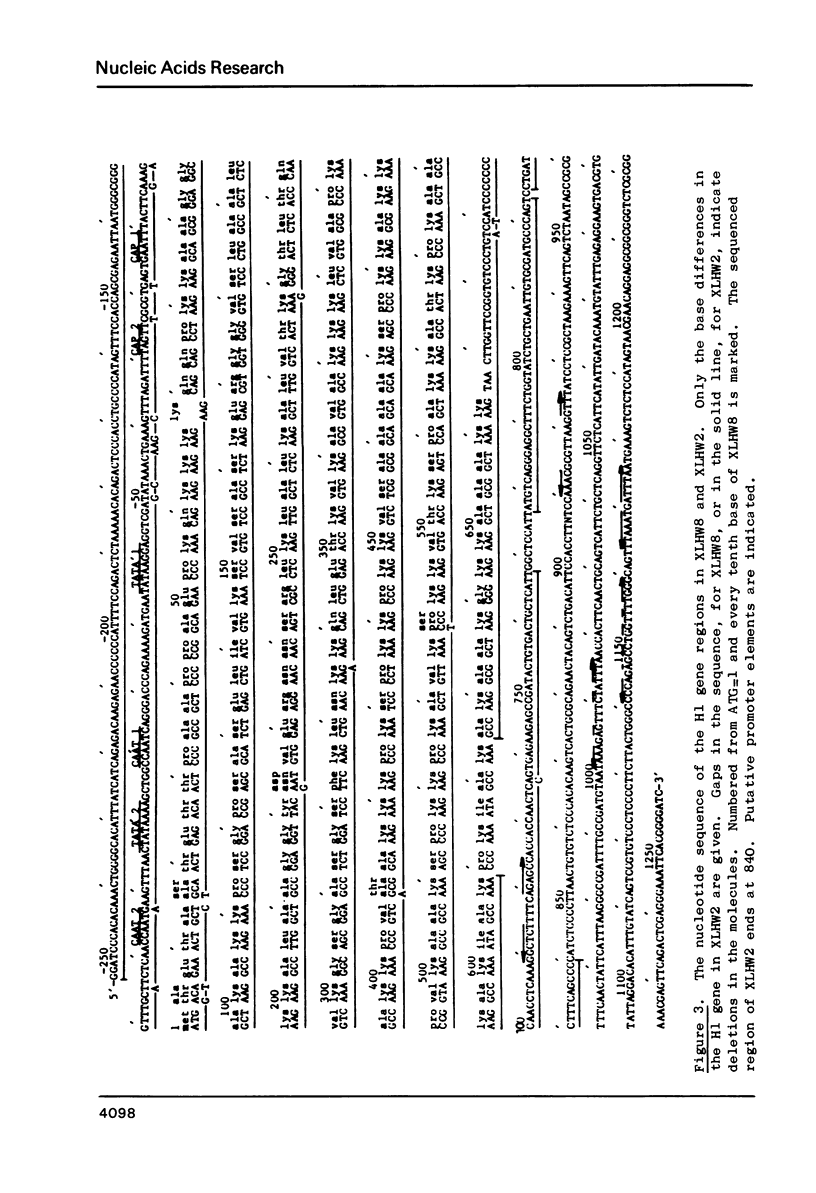
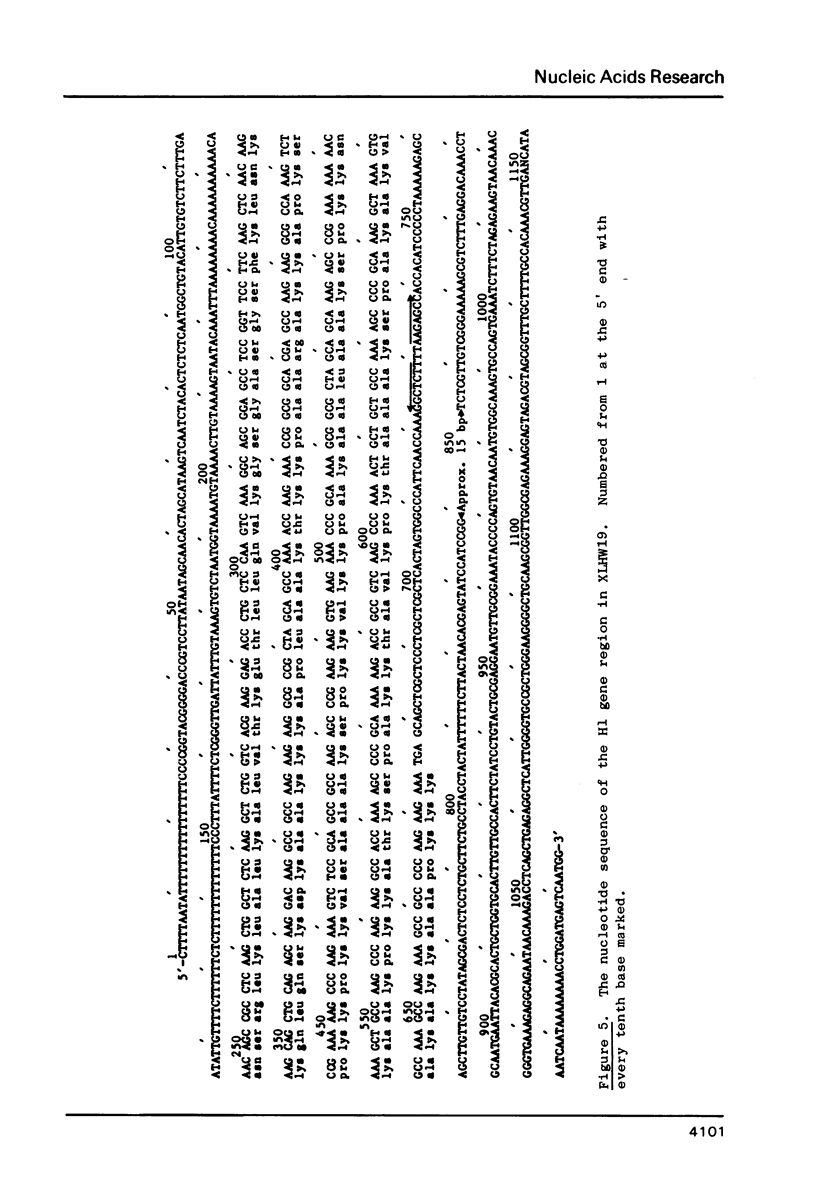
Four clones containing H1 histone gene sequences were previously isolated from a Xenopus laevis genomic library (1) and we now present the complete nucleotide sequences of these H1 genes and their flanking regions. Two of these genes code for minor H1 proteins, probably H1C, when expressed in the oocyte transcription/translation system and are present on clones with almost identical overall organization. However, at the nucleotide level these genes differ in showing base insertions and deletions, as well as substitutions. A third gene sequence which is more related to the major X. laevis H1A, corresponds to the 3' two thirds of an H1 gene. This gene has in place of a 5' coding region at least 1800 bp of apparently noncoding sequence, some of which is A-T rich. The junction does not correspond to the consensus sequence of an intron/exon boundary and therefore this H1 sequence is more likely to represent a pseudogene. Comparisons of the coding and flanking regions of these X. laevis H1 genes indicate the kind of differences which can occur among H1 subtypes within a species. A region of homology noted in the 3' noncoding portion of vertebrate histone genes is discussed in relation to the mechanism of termination of transcription.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birchmeier C., Grosschedl R., Birnstiel M. L. Generation of authentic 3' termini of an H2A mRNA in vivo is dependent on a short inverted DNA repeat and on spacer sequences. Cell. 1982 Apr;28(4):739–745. doi: 10.1016/0092-8674(82)90053-8. [DOI] [PubMed] [Google Scholar]
- Bustin M., Cole R. D. Regions of high and low cationic charge in a lysine-rich histone. J Biol Chem. 1970 Mar 25;245(6):1458–1466. [PubMed] [Google Scholar]
- Corden J., Wasylyk B., Buchwalder A., Sassone-Corsi P., Kedinger C., Chambon P. Promoter sequences of eukaryotic protein-coding genes. Science. 1980 Sep 19;209(4463):1406–1414. doi: 10.1126/science.6251548. [DOI] [PubMed] [Google Scholar]
- Destrée O. H., Hoenders H. J., Moorman A. F., Charles R. Histones of Xenopus laevis erythrocytes. Purification and characterization of the lysine-rich fractions. Biochim Biophys Acta. 1979 Mar 27;577(1):61–70. doi: 10.1016/0005-2795(79)90008-4. [DOI] [PubMed] [Google Scholar]
- Destrée O. H., d' Adelhart Toorop H. A., Charles R. Analysis of histones from different tissues and embryos of Xenopus laevis (Daudin). II. Qualitative and quantitative aspects of nuclear histones during early stages of development. Cell Differ. 1973 Oct;2(4):229–242. doi: 10.1016/0045-6039(73)90011-0. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A., Posakony J. W., Maniatis T., Lawn R. M., O'Connell C., Spritz R. A., DeRiel J. K., Forget B. G., Weissman S. M., Slightom J. L. The structure and evolution of the human beta-globin gene family. Cell. 1980 Oct;21(3):653–668. doi: 10.1016/0092-8674(80)90429-8. [DOI] [PubMed] [Google Scholar]
- Engel J. D., Sugarman B. J., Dodgson J. B. A chicken histone H3 gene contains intervening sequences. Nature. 1982 Jun 3;297(5865):434–436. doi: 10.1038/297434a0. [DOI] [PubMed] [Google Scholar]
- Flynn J. M., Woodland H. R. The synthesis of histone H1 during early amphibian development. Dev Biol. 1980 Mar;75(1):222–230. doi: 10.1016/0012-1606(80)90157-8. [DOI] [PubMed] [Google Scholar]
- Harvey R. P., Robins A. J., Wells J. R. Independently evolving chicken histone H2B genes: identification of a ubiquitous H2B-specific 5' element. Nucleic Acids Res. 1982 Dec 11;10(23):7851–7863. doi: 10.1093/nar/10.23.7851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel C. C., Birnstiel M. L. The organization and expression of histone gene families. Cell. 1981 Aug;25(2):301–313. doi: 10.1016/0092-8674(81)90048-9. [DOI] [PubMed] [Google Scholar]
- Isenberg I. Histones. Annu Rev Biochem. 1979;48:159–191. doi: 10.1146/annurev.bi.48.070179.001111. [DOI] [PubMed] [Google Scholar]
- Krieg P. A., Robins A. J., Colman A., Wells J. R. Chicken histone H5 mRNA: the polyadenylated RNA lacks the conserved histone 3' terminator sequence. Nucleic Acids Res. 1982 Nov 11;10(21):6777–6785. doi: 10.1093/nar/10.21.6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S., Sures I., Kedes L. The nucleotide and amino acid coding sequence of a gene for H1 histone that interacts with euchromatin. The early embryonic H1 gene of the sea urchin Strongylocentrotus purpuratus. J Biol Chem. 1982 Aug 25;257(16):9438–9443. [PubMed] [Google Scholar]
- Macleod A. R., Wong N. C., Dixon G. H. The amino-acid sequence of trout-testis histone H1. Eur J Biochem. 1977 Aug 15;78(1):281–291. doi: 10.1111/j.1432-1033.1977.tb11739.x. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Moorman A. F., De Boer P. A., De Laaf R. T., Destrée O. H. Primary structure of the histone H2A and H2B genes and their flanking sequences in a minor histone gene cluster of Xenopus laevis. FEBS Lett. 1982 Aug 2;144(2):235–241. doi: 10.1016/0014-5793(82)80645-5. [DOI] [PubMed] [Google Scholar]
- Moorman A. F., de Boer P. A., de Laaf R. T., van Dongen W. M., Destrée O. H. Primary structure of the histone H3 and H4 genes and their flanking sequences in a minor histone gene cluster of Xenopus laevis. FEBS Lett. 1981 Dec 21;136(1):45–52. doi: 10.1016/0014-5793(81)81211-2. [DOI] [PubMed] [Google Scholar]
- Morris N. R. A comparison of the structure of chicken erythrocyte and chicken liver chromatin. Cell. 1976 Dec;9(4 Pt 1):627–632. doi: 10.1016/0092-8674(76)90045-3. [DOI] [PubMed] [Google Scholar]
- Newrock K. M., Cohen L. H., Hendricks M. B., Donnelly R. J., Weinberg E. S. Stage-specific mRNAs coding for subtypes of H2A and H2B histones in the sea urchin embryo. Cell. 1978 Jun;14(2):327–336. doi: 10.1016/0092-8674(78)90118-6. [DOI] [PubMed] [Google Scholar]
- Noll M. Differences and similarities in chromatin structure of Neurospora crassa and higher eucaryotes. Cell. 1976 Jul;8(3):349–355. doi: 10.1016/0092-8674(76)90146-x. [DOI] [PubMed] [Google Scholar]
- Old R. W., Woodland H. R., Ballantine J. E., Aldridge T. C., Newton C. A., Bains W. A., Turner P. C. Organization and expression of cloned histone gene clusters from Xenopus laevis and X. borealis. Nucleic Acids Res. 1982 Dec 11;10(23):7561–7580. doi: 10.1093/nar/10.23.7561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehrson J. R., Cole R. D. Bovine H10 histone subfractions contain an invariant sequence which matches histones H5 rather than H1. Biochemistry. 1981 Apr 14;20(8):2298–2301. doi: 10.1021/bi00511a035. [DOI] [PubMed] [Google Scholar]
- Risley M. S., Eckhardt R. A. H1 histone variants in Xenopus laevis. Dev Biol. 1981 May;84(1):79–87. doi: 10.1016/0012-1606(81)90372-9. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner W., Kunz G., Daetwyler H., Telford J., Smith H. O., Birnstiel M. L. Genes and spacers of cloned sea urchin histone DNA analyzed by sequencing. Cell. 1978 Jul;14(3):655–671. doi: 10.1016/0092-8674(78)90249-0. [DOI] [PubMed] [Google Scholar]
- Seiler-Tuyns A., Birnstiel M. L. Structure and expression in L-cells of a cloned H4 histone gene of the mouse. J Mol Biol. 1981 Oct 5;151(4):607–625. doi: 10.1016/0022-2836(81)90426-5. [DOI] [PubMed] [Google Scholar]
- Sharp P. A. Speculations on RNA splicing. Cell. 1981 Mar;23(3):643–646. doi: 10.1016/0092-8674(81)90425-6. [DOI] [PubMed] [Google Scholar]
- Stunnenberg H. G., Birnstiel M. L. Bioassay for components regulating eukaryotic gene expression: a chromosomal factor involved in the generation of histone mRNA 3' termini. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6201–6204. doi: 10.1073/pnas.79.20.6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner P. C., Woodland H. R. H3 and H4 histone cDNA sequences from Xenopus: a sequence comparison of H4 genes. Nucleic Acids Res. 1982 Jun 25;10(12):3769–3780. doi: 10.1093/nar/10.12.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner P. C., Woodland H. R. Histone gene number and organisation in Xenopus: Xenopus borealis has a homogeneous major cluster. Nucleic Acids Res. 1983 Feb 25;11(4):971–986. doi: 10.1093/nar/11.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Holt C., Strickland W. N., Brandt W. F., Strickland M. S. More histone structures. FEBS Lett. 1979 Apr 15;100(2):201–218. doi: 10.1016/0014-5793(79)80337-3. [DOI] [PubMed] [Google Scholar]
- Weintraub H. The nucleosome repeat length increases during erythropoiesis in the chick. Nucleic Acids Res. 1978 Apr;5(4):1179–1188. doi: 10.1093/nar/5.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaguchi M., Roy C., Seligy V. L. Complete amino acid sequence of goose erythrocyte H5 histone and the homology between H1 and H5 histones. Biochem Biophys Res Commun. 1979 Oct 29;90(4):1400–1406. doi: 10.1016/0006-291x(79)91191-4. [DOI] [PubMed] [Google Scholar]
- Zernik M., Heintz N., Boime I., Roeder R. G. Xenopus laevis histone genes: variant H1 genes are present in different clusters. Cell. 1980 Dec;22(3):807–815. doi: 10.1016/0092-8674(80)90557-7. [DOI] [PubMed] [Google Scholar]



