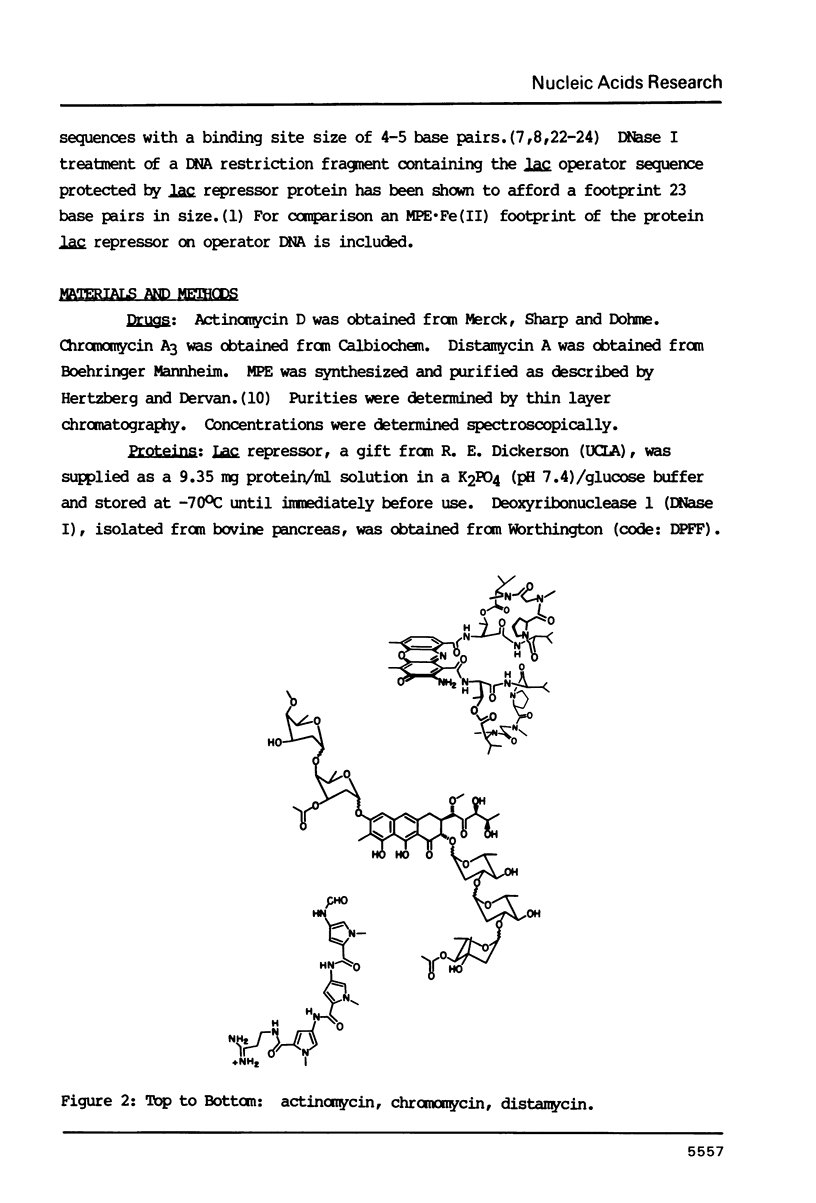
Abstract
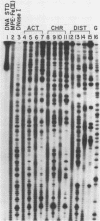
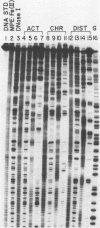
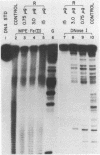
DNase I and MPE.Fe (II) footprinting both employ partial cleavage of ligand-protected DNA restriction fragments and Maxam-Gilbert sequencing gel methods of analysis. One method utilizes the enzyme, DNase I, as the DNA cleaving agent while the other employs the synthetic molecule, methidium-propyl-EDTA (MPE). For actinomycin D, chromomycin A3 and distamycin A, DNase I footprinting reports larger binding site sizes than MPE.Fe (II). DNase I footprinting appears more sensitive for weakly bound sites. MPE.Fe (II) footprinting appears more accurate in determining the actual size and location of the binding sites for small molecules on DNA, especially in cases where several small molecules are closely spaced on the DNA. MPE.Fe (II) and DNase I report the same sequence and binding site size for lac repressor protein on operator DNA.
Full text
PDF




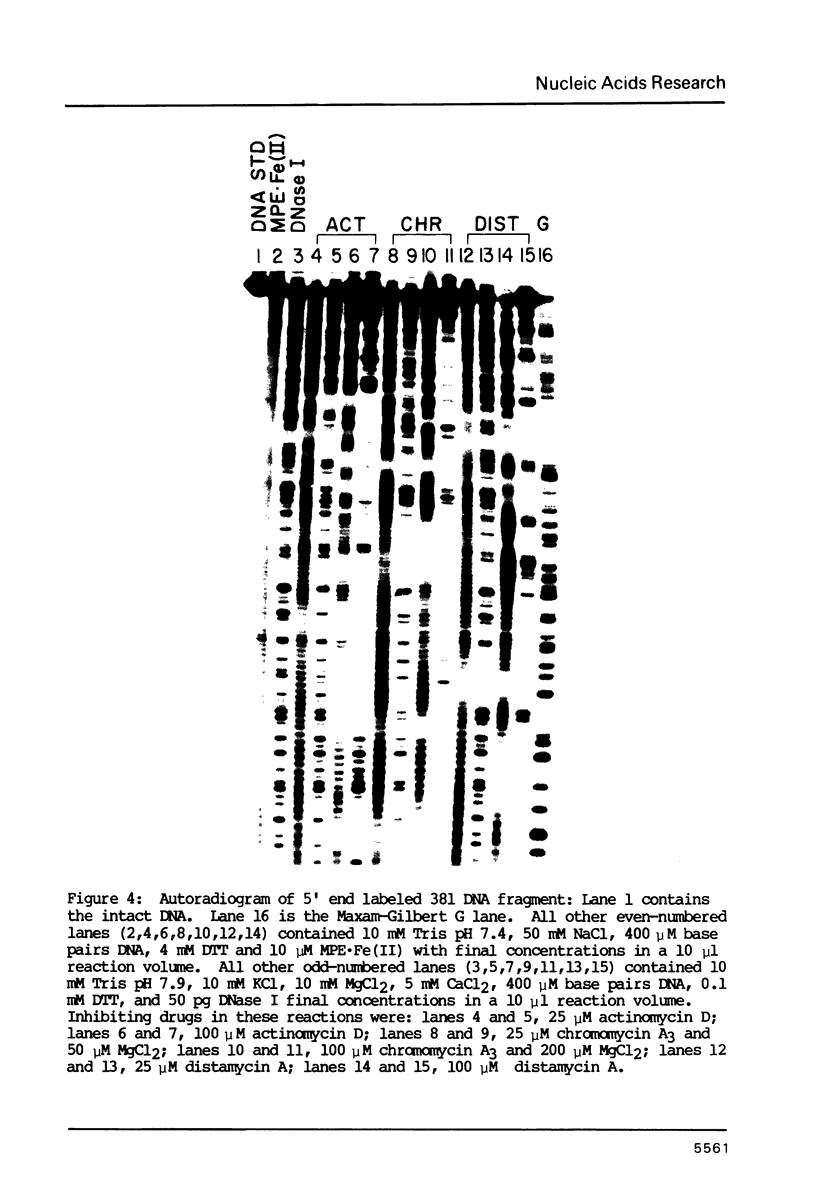






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Behr W., Honikel K., Hartmann G. Interaction of the RNA polymerase inhibitor chromomycin with DNA. Eur J Biochem. 1969 May 1;9(1):82–92. doi: 10.1111/j.1432-1033.1969.tb00579.x. [DOI] [PubMed] [Google Scholar]
- D'Andrea A. D., Haseltine W. A. Sequence specific cleavage of DNA by the antitumor antibiotics neocarzinostatin and bleomycin. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3608–3612. doi: 10.1073/pnas.75.8.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOLDBERG I. H., RABINOWITZ M., REICH E. Basis of actinomycin action. I. DNA binding and inhibition of RNA-polymerase synthetic reactions by actinomycin. Proc Natl Acad Sci U S A. 1962 Dec 15;48:2094–2101. doi: 10.1073/pnas.48.12.2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseltine W. A., Lindan C. P., D'Andrea A. D., Johnsrud L. The use of DNA fragments of defined sequence for the study of DNA damage and repair. Methods Enzymol. 1980;65(1):235–248. doi: 10.1016/s0076-6879(80)65033-2. [DOI] [PubMed] [Google Scholar]
- Kross J., Henner W. D., Hecht S. M., Haseltine W. A. Specificity of deoxyribonucleic acid cleavage by bleomycin, phleomycin, and tallysomycin. Biochemistry. 1982 Aug 31;21(18):4310–4318. doi: 10.1021/bi00261a021. [DOI] [PubMed] [Google Scholar]
- Krylov A. S., Grokhovsky S. L., Zasedatelev A. S., Zhuze A. L., Gursky G. V., Gottikh B. P. Quantitative estimation of the contribution of pyrrolcarboxamide groups of the antibiotic distamycin A into specificity of its binding to DNA AT pairs. Nucleic Acids Res. 1979 Jan;6(1):289–304. doi: 10.1093/nar/6.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane M. J., Dabrowiak J. C., Vournakis J. N. Sequence specificity of actinomycin D and Netropsin binding to pBR322 DNA analyzed by protection from DNase I. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3260–3264. doi: 10.1073/pnas.80.11.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonossoff G. P., Butler P. J., Klug A. Sequence-dependent variation in the conformation of DNA. J Mol Biol. 1981 Jul 15;149(4):745–760. doi: 10.1016/0022-2836(81)90356-9. [DOI] [PubMed] [Google Scholar]
- Luck G., Triebel H., Waring M., Zimmer C. Conformation dependent binding of netropsin and distamycin to DNA and DNA model polymers. Nucleic Acids Res. 1974 Mar;1(3):503–530. doi: 10.1093/nar/1.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Müller W., Crothers D. M. Studies of the binding of actinomycin and related compounds to DNA. J Mol Biol. 1968 Jul 28;35(2):251–290. doi: 10.1016/s0022-2836(68)80024-5. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Rice J. A., Broka C., Itakura K. Mutual interaction between adjacent dG . dC actinomycin binding sites and dA . dT netropsin binding sites on the self-complementary d(C-G-C-G-A-A-T-T-C-G-C-G) duplex in solution. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7281–7284. doi: 10.1073/pnas.78.12.7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobell H. M. The stereochemistry of actinomycin binding to DNA and its implications in molecular biology. Prog Nucleic Acid Res Mol Biol. 1973;13:153–190. doi: 10.1016/s0079-6603(08)60103-8. [DOI] [PubMed] [Google Scholar]
- Takeshita M., Grollman A. P., Ohtsubo E., Ohtsubo H. Interaction of bleomycin with DNA. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5983–5987. doi: 10.1073/pnas.75.12.5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takusagawa F., Dabrow M., Neidle S., Berman H. M. The structure of a pseudo intercalated complex between actinomycin and the DNA binding sequence d(GpC). Nature. 1982 Apr 1;296(5856):466–469. doi: 10.1038/296466a0. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Dervan P. B. Chromomycin, mithramycin, and olivomycin binding sites on heterogeneous deoxyribonucleic acid. Footprinting with (methidiumpropyl-EDTA)iron(II). Biochemistry. 1983 May 10;22(10):2373–2377. doi: 10.1021/bi00279a011. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Dervan P. B. Footprinting with MPE.Fe(II). Complementary-strand analyses of distamycin- and actinomycin-binding sites on heterogeneous DNA. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):347–353. doi: 10.1101/sqb.1983.047.01.040. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Hertzberg R. P., Dervan P. B. Map of distamycin, netropsin, and actinomycin binding sites on heterogeneous DNA: DNA cleavage-inhibition patterns with methidiumpropyl-EDTA.Fe(II). Proc Natl Acad Sci U S A. 1982 Sep;79(18):5470–5474. doi: 10.1073/pnas.79.18.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]