Abstract
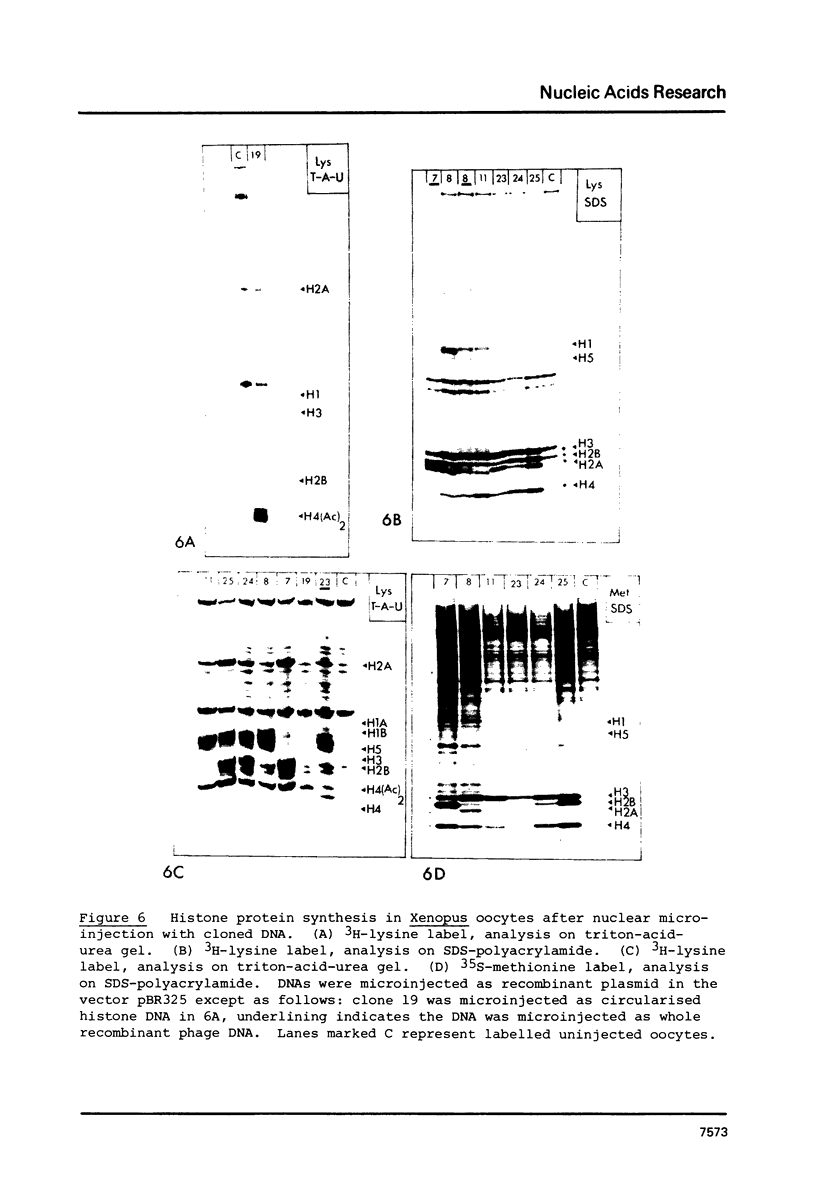
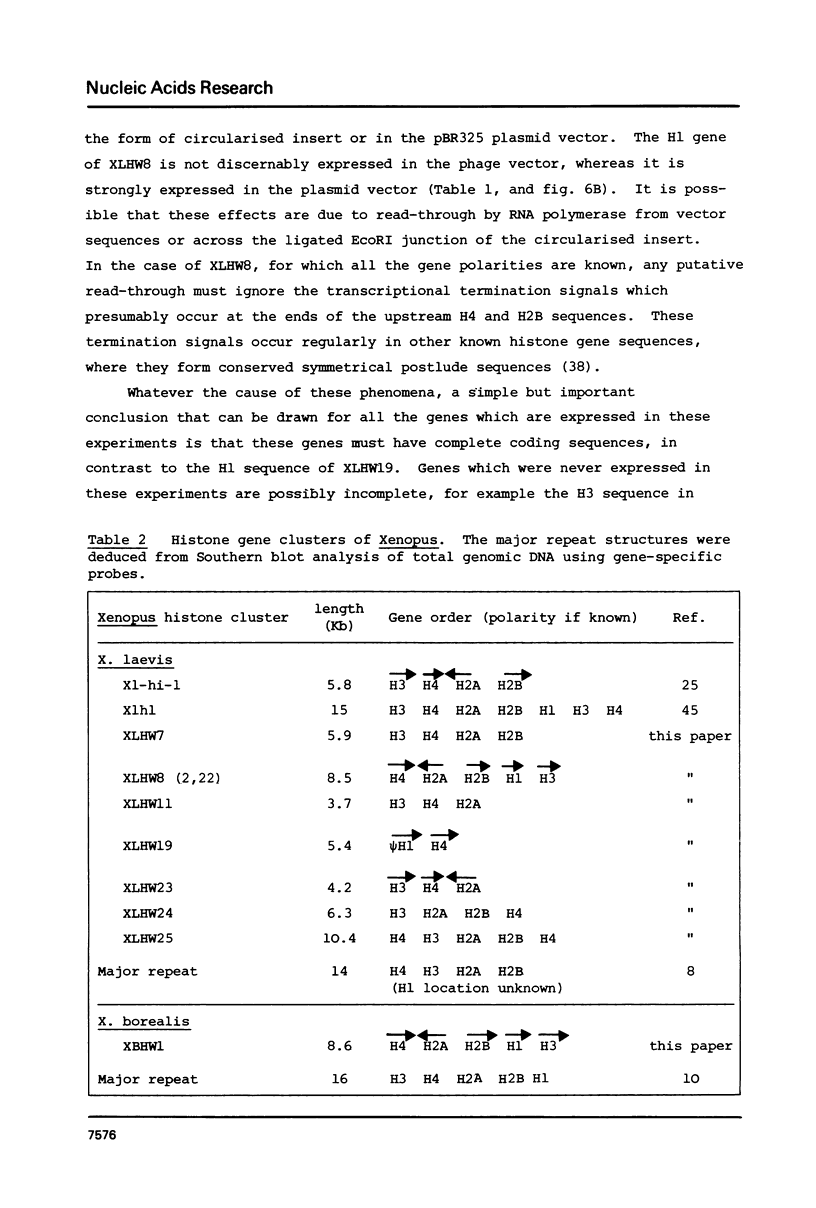
We have isolated several clones containing Xenopus histone genes from genomic libraries of X. laevis and X. borealis DNA. Each genomic clone has been mapped and the positions of 26 histone genes in seven laevis clones and 5 histone genes in one borealis clone have been determined. In laevis, the histone gene clusters show considerable variation in gene order within a single individual. When the cloned DNAs were microinjected into the nucleus of Xenopus oocytes, expression of cloned genes at the transcriptional and translational level was readily detectable. Previously unknown histone variants were revealed by the microinjection experiments.
Full text
PDF







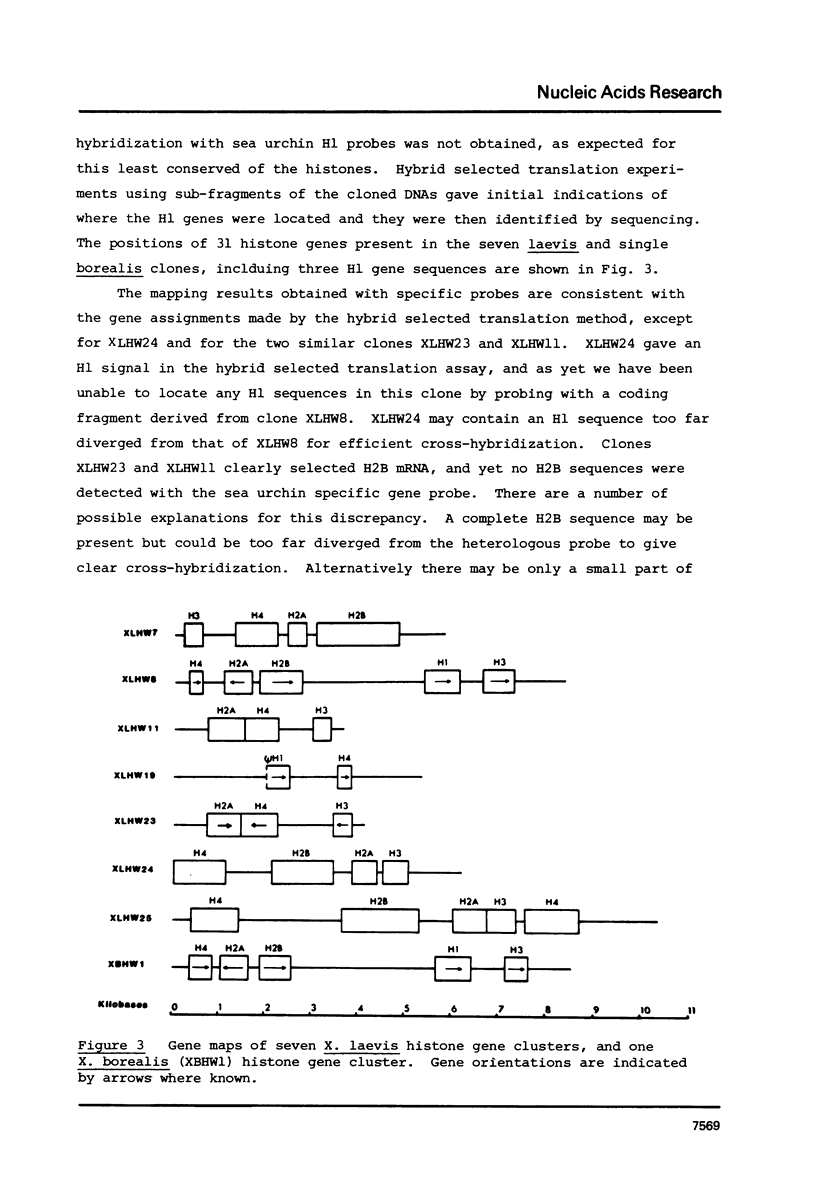









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adamson E. D., Woodland H. R. Histone synthesis in early amphibian development: histone and DNA syntheses are not co-ordinated. J Mol Biol. 1974 Sep 15;88(2):263–285. doi: 10.1016/0022-2836(74)90481-1. [DOI] [PubMed] [Google Scholar]
- Albright S. C., Nelson P. P., Garrard W. T. Histone molar ratios among different electrophoretic forms of mono- and dinucleosomes. J Biol Chem. 1979 Feb 25;254(4):1065–1073. [PubMed] [Google Scholar]
- Alfageme C. R., Zweidler A., Mahowald A., Cohen L. H. Histones of Drosophila embryos. Electrophoretic isolation and structural studies. J Biol Chem. 1974 Jun 25;249(12):3729–3736. [PubMed] [Google Scholar]
- Arceci R. J., Gross P. R. Noncoincidence of histone and DNA synthesis in cleavage cycles of early development. Proc Natl Acad Sci U S A. 1977 Nov;74(11):5016–5020. doi: 10.1073/pnas.74.11.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARTH L. G., BARTH L. J. Differentiation of cells of the Rana pipiens gastrula in unconditioned medium. J Embryol Exp Morphol. 1959 Jun;7:210–222. [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Birchmeier C., Grosschedl R., Birnstiel M. L. Generation of authentic 3' termini of an H2A mRNA in vivo is dependent on a short inverted DNA repeat and on spacer sequences. Cell. 1982 Apr;28(4):739–745. doi: 10.1016/0092-8674(82)90053-8. [DOI] [PubMed] [Google Scholar]
- Bisbee C. A., Baker M. A., Wilson A. C., Haji-Azimi I., Fischberg M. Albumin phylogeny for clawed frogs (Xenopus). Science. 1977 Feb 25;195(4280):785–787. doi: 10.1126/science.65013. [DOI] [PubMed] [Google Scholar]
- Blattner F. R., Williams B. G., Blechl A. E., Denniston-Thompson K., Faber H. E., Furlong L., Grunwald D. J., Kiefer D. O., Moore D. D., Schumm J. W. Charon phages: safer derivatives of bacteriophage lambda for DNA cloning. Science. 1977 Apr 8;196(4286):161–169. doi: 10.1126/science.847462. [DOI] [PubMed] [Google Scholar]
- Bolivar F. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique Eco RI sites for selection of Eco RI generated recombinant DNA molecules. Gene. 1978 Oct;4(2):121–136. doi: 10.1016/0378-1119(78)90025-2. [DOI] [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Destrée O. H., Hoenders H. J., Moorman A. F., Charles R. Histones of Xenopus laevis erythrocytes. Purification and characterization of the lysine-rich fractions. Biochim Biophys Acta. 1979 Mar 27;577(1):61–70. doi: 10.1016/0005-2795(79)90008-4. [DOI] [PubMed] [Google Scholar]
- Engel J. D., Dodgson J. B. Histone genes are clustered but not tandemly repeated in the chicken genome. Proc Natl Acad Sci U S A. 1981 May;78(5):2856–2860. doi: 10.1073/pnas.78.5.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquist L., Sternberg N. In vitro packaging of lambda Dam vectors and their use in cloning DNA fragments. Methods Enzymol. 1979;68:281–298. doi: 10.1016/0076-6879(79)68020-5. [DOI] [PubMed] [Google Scholar]
- Heintz N., Zernik M., Roeder R. G. The structure of the human histone genes: clustered but not tandemly repeated. Cell. 1981 Jun;24(3):661–668. doi: 10.1016/0092-8674(81)90092-1. [DOI] [PubMed] [Google Scholar]
- Hentschel C. C., Birnstiel M. L. The organization and expression of histone gene families. Cell. 1981 Aug;25(2):301–313. doi: 10.1016/0092-8674(81)90048-9. [DOI] [PubMed] [Google Scholar]
- Hentschel C., Probst E., Birnstiel M. L. Transcriptional fidelity of histone genes injected into Xenopus oocyte nuclei. Nature. 1980 Nov 6;288(5786):100–102. doi: 10.1038/288100a0. [DOI] [PubMed] [Google Scholar]
- Isenberg I. Histones. Annu Rev Biochem. 1979;48:159–191. doi: 10.1146/annurev.bi.48.070179.001111. [DOI] [PubMed] [Google Scholar]
- KIRBY K. S. ISOLATION AND CHARACTERIZATION OF RIBOSOMAL RIBONUCLEIC ACID. Biochem J. 1965 Jul;96:266–269. doi: 10.1042/bj0960266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder P., Tiemeier D., Enquist L. EK2 derivatives of bacteriophage lambda useful in the cloning of DNA from higher organisms: the lambdagtWES system. Science. 1977 Apr 8;196(4286):175–177. doi: 10.1126/science.322278. [DOI] [PubMed] [Google Scholar]
- Levy S., Childs G., Kedes L. Sea urchin nuclei use RNA polymerase II to transcribe discrete histone RNAs larger than messengers. Cell. 1978 Sep;15(1):151–162. doi: 10.1016/0092-8674(78)90091-0. [DOI] [PubMed] [Google Scholar]
- Mertz J. E., Gurdon J. B. Purified DNAs are transcribed after microinjection into Xenopus oocytes. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1502–1506. doi: 10.1073/pnas.74.4.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman A. F., de Laaf R. T., Destrée O. H., Telford J., Birnstiel M. L. Histone genes from Xenopus laevis: molecular cloning and initial characterization. Gene. 1980 Aug;10(3):185–193. doi: 10.1016/0378-1119(80)90048-7. [DOI] [PubMed] [Google Scholar]
- Newrock K. M., Alfageme C. R., Nardi R. V., Cohen L. H. Histone changes during chromatin remodeling in embryogenesis. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):421–431. doi: 10.1101/sqb.1978.042.01.045. [DOI] [PubMed] [Google Scholar]
- O'Farrell P. H., Kutter E., Nakanishi M. A restriction map of the bacteriophage T4 genome. Mol Gen Genet. 1980;179(2):421–435. doi: 10.1007/BF00425473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst E., Kressmann A., Birnstiel M. L. Expression of sea urchin histone genes in the oocyte of Xenopus laevis. J Mol Biol. 1979 Dec 15;135(3):709–732. doi: 10.1016/0022-2836(79)90173-6. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Risley M. S., Eckhardt R. A. H1 histone variants in Xenopus laevis. Dev Biol. 1981 May;84(1):79–87. doi: 10.1016/0012-1606(81)90372-9. [DOI] [PubMed] [Google Scholar]
- Risley M. S., Eckhardt R. A., Mann M., Kasinsky H. E. Determinants of sperm nuclear shaping in the genus Xenopus. Chromosoma. 1982;84(4):557–569. doi: 10.1007/BF00292855. [DOI] [PubMed] [Google Scholar]
- Roberts B. E., Paterson B. M. Efficient translation of tobacco mosaic virus RNA and rabbit globin 9S RNA in a cell-free system from commercial wheat germ. Proc Natl Acad Sci U S A. 1973 Aug;70(8):2330–2334. doi: 10.1073/pnas.70.8.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra F., Lichtler A., Marashi F., Rickles R., Van Dyke T., Clark S., Wells J., Stein G., Stein J. Organization of human histone genes. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1795–1799. doi: 10.1073/pnas.79.6.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittman D. B., Chiu I. M., Pan C. J., Cohn R. H., Kedes L. H., Marzluff W. F. Isolation of two clusters of mouse histone genes. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4078–4082. doi: 10.1073/pnas.78.7.4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stephenson E. C., Erba H. P., Gall J. G. Characterization of a cloned histone gene cluster of the newt Notophthalamus viridescens. Nucleic Acids Res. 1981 May 25;9(10):2281–2295. doi: 10.1093/nar/9.10.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sures I., Lowry J., Kedes L. H. The DNA sequence of sea urchin (S. purpuratus) H2A, H2B and H3 histone coding and spacer regions. Cell. 1978 Nov;15(3):1033–1044. doi: 10.1016/0092-8674(78)90287-8. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Kornberg R. D. Cleavable cross-links in the analysis of histone-histone associations. FEBS Lett. 1975 Oct 15;58(1):353–358. doi: 10.1016/0014-5793(75)80296-1. [DOI] [PubMed] [Google Scholar]
- Turner P. C., Woodland H. R. H3 and H4 histone cDNA sequences from Xenopus: a sequence comparison of H4 genes. Nucleic Acids Res. 1982 Jun 25;10(12):3769–3780. doi: 10.1093/nar/10.12.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodland H. R., Adamson E. D. The synthesis and storage of histones during the oogenesis of Xenopus laevis. Dev Biol. 1977 May;57(1):118–135. doi: 10.1016/0012-1606(77)90359-1. [DOI] [PubMed] [Google Scholar]
- Woodland H. R. The modification of stored histones H3 and H4 during the oogenesis and early development of Xenopus laevis. Dev Biol. 1979 Feb;68(2):360–370. doi: 10.1016/0012-1606(79)90210-0. [DOI] [PubMed] [Google Scholar]
- Wu R. S., Bonner W. M. Separation of basal histone synthesis from S-phase histone synthesis in dividing cells. Cell. 1981 Dec;27(2 Pt 1):321–330. doi: 10.1016/0092-8674(81)90415-3. [DOI] [PubMed] [Google Scholar]
- Zernik M., Heintz N., Boime I., Roeder R. G. Xenopus laevis histone genes: variant H1 genes are present in different clusters. Cell. 1980 Dec;22(3):807–815. doi: 10.1016/0092-8674(80)90557-7. [DOI] [PubMed] [Google Scholar]
- van Dongen W., de Laaf L., Zaal R., Moorman A., Destrée O. The organization of the histone genes in the genome of Xenopus laevis. Nucleic Acids Res. 1981 May 25;9(10):2297–2311. doi: 10.1093/nar/9.10.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]