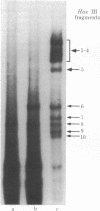
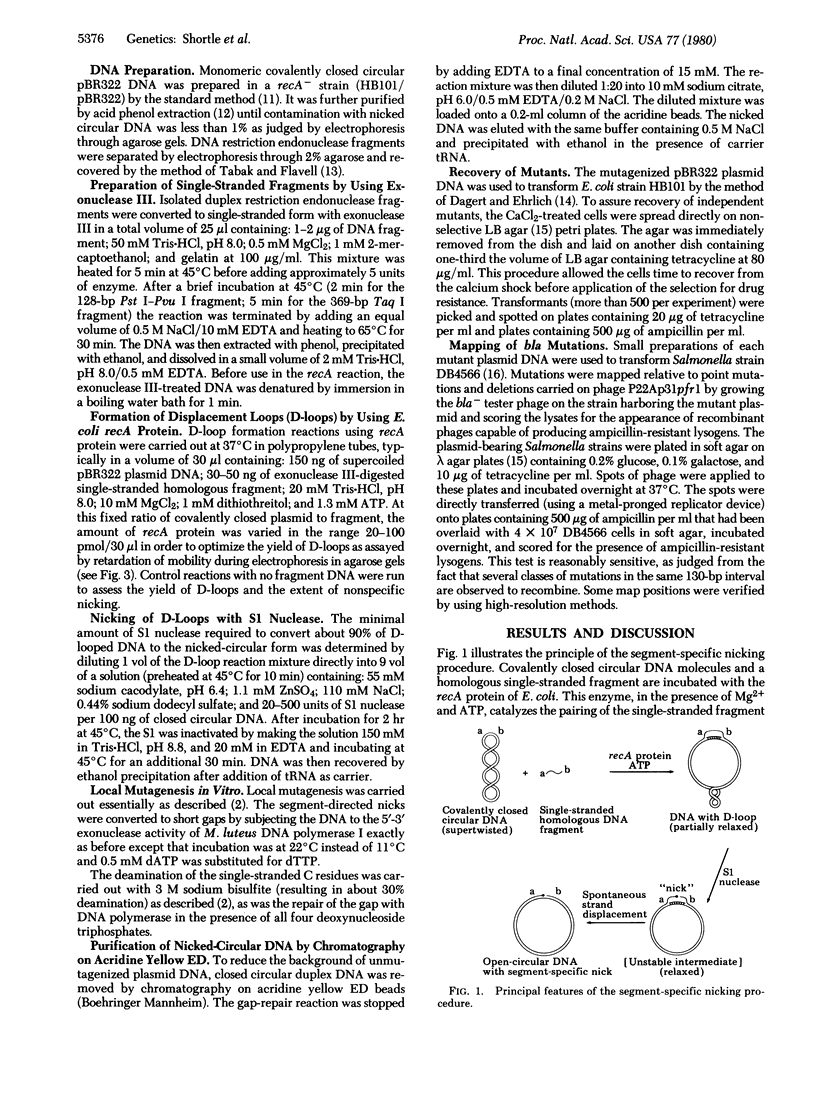
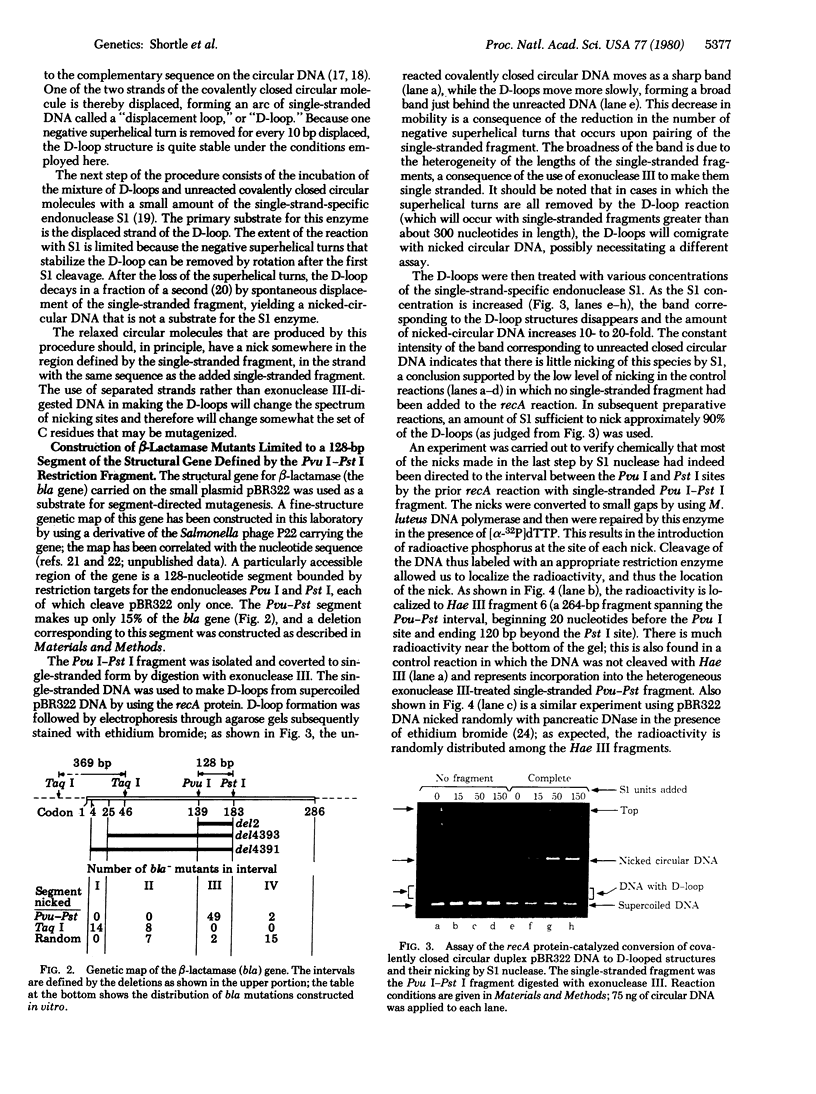
Abstract
A general method for efficiently mutagenizing a predetermined segment of a closed circular duplex DNA molecule was used to construct mutations in two specific regions of the beta-lactamase (bla) gene carried by the small plasmid pBR322. The principle of segment-directed mutagenesis is the use of a single-stranded homologous DNA fragment to direct the nicking of circular duplex DNA within a segment defined by the DNA fragment in a two-step reaction. First, Escherichia coli recA protein is used to catalyze assimilation of the homologous single-stranded DNA, producing a displacement loop ("D-loop") in the circular DNA. Second, a small amount of the single-strand-specific S1 nuclease is used to nick the displaced DNA. The segment-directed nicks are converted to small gaps, which are then mutagenized specifically with sodium bisulfite. A short (128-base pair) restriction endonuclease fragment from the center of the bla gene was used to direct mutagenesis with the result that 7.5% of the recovered plasmids were bla- mutants and 49/51 of these mutants, mapped genetically, were found to lie in a deletion interval whose endpoints approximate those of the restriction fragment. Similar results were obtained when another short fragment covering the beginning of the gene was used; many of these mutations map in the region coding the "signal" sequence thought to be involved in secretion of beta-lactamase.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambler R. P., Scott G. K. Partial amino acid sequence of penicillinase coded by Escherichia coli plasmid R6K. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3732–3736. doi: 10.1073/pnas.75.8.3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrias W. E., Wilschut I. J., Vereijken J. M., Weisbeek P. J., van Arkel G. A. Induction and isolation of mutants in a specific region of gene A of bacteriophage phi chi 174. Virology. 1976 Mar;70(1):195–197. doi: 10.1016/0042-6822(76)90251-8. [DOI] [PubMed] [Google Scholar]
- Chu C. T., Parris D. S., Dixon R. A., Farber F. E., Schaffer P. A. Hydroxylamine mutagenesis of HSV DNA and DNA fragments: introduction of mutations into selected regions of the viral genome. Virology. 1979 Oct 15;98(1):168–181. doi: 10.1016/0042-6822(79)90535-x. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979 May;6(1):23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- DiMaio D., Nathans D. Cold-sensitive regulatory mutants of simian virus 40. J Mol Biol. 1980 Jun 15;140(1):129–142. doi: 10.1016/0022-2836(80)90359-9. [DOI] [PubMed] [Google Scholar]
- Green C., Tibbetts C. Targeted deletions of sequences from closed circular DNA. Proc Natl Acad Sci U S A. 1980 May;77(5):2455–2459. doi: 10.1073/pnas.77.5.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield L., Simpson L., Kaplan D. Conversion of closed circular DNA molecules to single-nicked molecules by digestion with DNAase I in the presence of ethidium bromide. Biochim Biophys Acta. 1975 Oct 15;407(3):365–375. doi: 10.1016/0005-2787(75)90104-5. [DOI] [PubMed] [Google Scholar]
- Hutchison C. A., 3rd, Phillips S., Edgell M. H., Gillam S., Jahnke P., Smith M. Mutagenesis at a specific position in a DNA sequence. J Biol Chem. 1978 Sep 25;253(18):6551–6560. [PubMed] [Google Scholar]
- Koshland D., Botstein D. Secretion of beta-lactamase requires the carboxy end of the protein. Cell. 1980 Jul;20(3):749–760. doi: 10.1016/0092-8674(80)90321-9. [DOI] [PubMed] [Google Scholar]
- Lederberg E. M., Cohen S. N. Transformation of Salmonella typhimurium by plasmid deoxyribonucleic acid. J Bacteriol. 1974 Sep;119(3):1072–1074. doi: 10.1128/jb.119.3.1072-1074.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEntee K., Weinstock G. M., Lehman I. R. Initiation of general recombination catalyzed in vitro by the recA protein of Escherichia coli. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2615–2619. doi: 10.1073/pnas.76.6.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller W., Weber H., Meyer F., Weissmann C. Site-directed mutagenesis in DNA: generation of point mutations in cloned beta globin complementary dna at the positions corresponding to amino acids 121 to 123. J Mol Biol. 1978 Sep 15;124(2):343–358. doi: 10.1016/0022-2836(78)90303-0. [DOI] [PubMed] [Google Scholar]
- Parker R. C., Watson R. M., Vinograd J. Mapping of closed circular DNAs by cleavage with restriction endonucleases and calibration by agarose gel electrophoresis. Proc Natl Acad Sci U S A. 1977 Mar;74(3):851–855. doi: 10.1073/pnas.74.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock A. C., Dingman C. W. Resolution of multiple ribonucleic acid species by polyacrylamide gel electrophoresis. Biochemistry. 1967 Jun;6(6):1818–1827. doi: 10.1021/bi00858a033. [DOI] [PubMed] [Google Scholar]
- Radding C. M., Beattie K. L., Holloman W. K., Wiegand R. C. Uptake of homologous single-stranded fragments by superhelical DNA. IV. Branch migration. J Mol Biol. 1977 Nov;116(4):825–839. doi: 10.1016/0022-2836(77)90273-x. [DOI] [PubMed] [Google Scholar]
- Shibata T., DasGupta C., Cunningham R. P., Radding C. M. Purified Escherichia coli recA protein catalyzes homologous pairing of superhelical DNA and single-stranded fragments. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1638–1642. doi: 10.1073/pnas.76.4.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortle D., Nathans D. Local mutagenesis: a method for generating viral mutants with base substitutions in preselected regions of the viral genome. Proc Natl Acad Sci U S A. 1978 May;75(5):2170–2174. doi: 10.1073/pnas.75.5.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabak H. F., Flavell R. A. A method for the recovery of DNA from agarose gels. Nucleic Acids Res. 1978 Jul;5(7):2321–2332. doi: 10.1093/nar/5.7.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt V. M. Purification and further properties of single-strand-specific nuclease from Aspergillus oryzae. Eur J Biochem. 1973 Feb 15;33(1):192–200. doi: 10.1111/j.1432-1033.1973.tb02669.x. [DOI] [PubMed] [Google Scholar]
- Weinstock G. M., McEntee K., Lehman I. R. ATP-dependent renaturation of DNA catalyzed by the recA protein of Escherichia coli. Proc Natl Acad Sci U S A. 1979 Jan;76(1):126–130. doi: 10.1073/pnas.76.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasloff M., Ginder G. D., Felsenfeld G. A new method for the purification and identification of covalently closed circular DNA molcules. Nucleic Acids Res. 1978 Apr;5(4):1139–1152. doi: 10.1093/nar/5.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]