Abstract
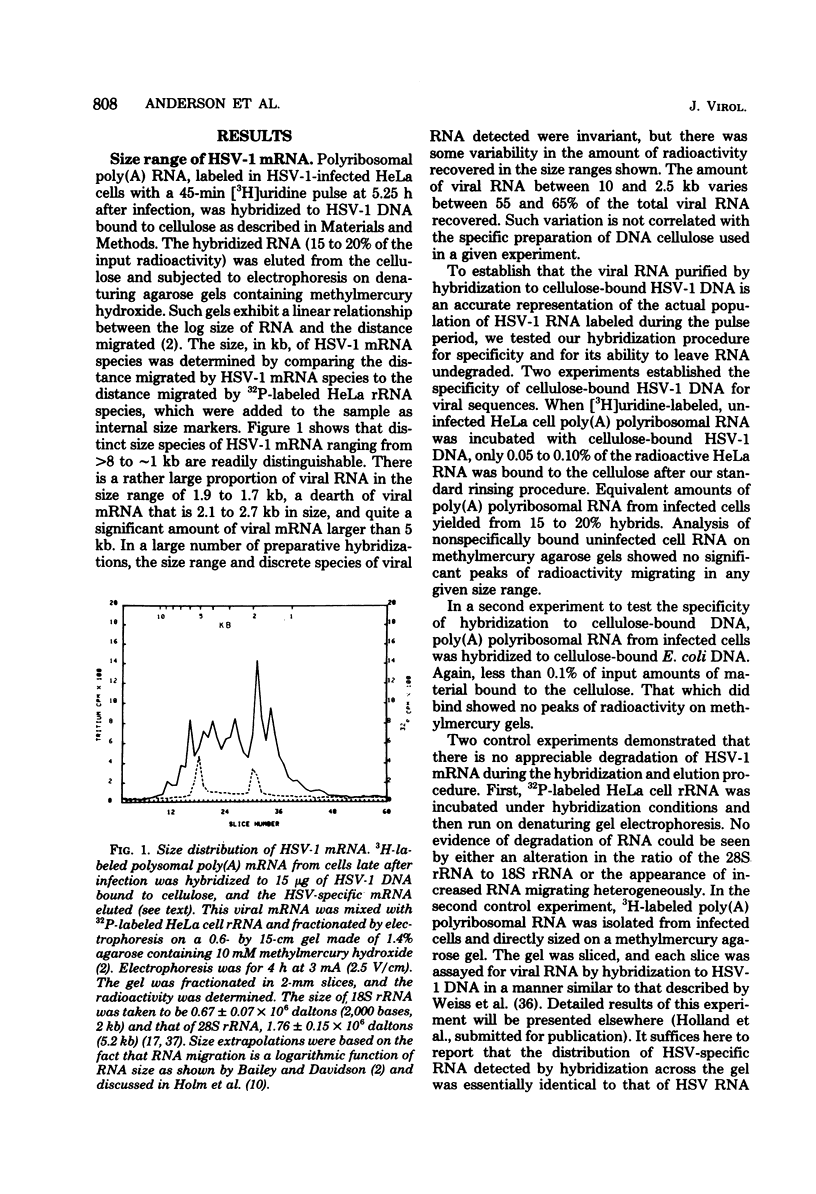
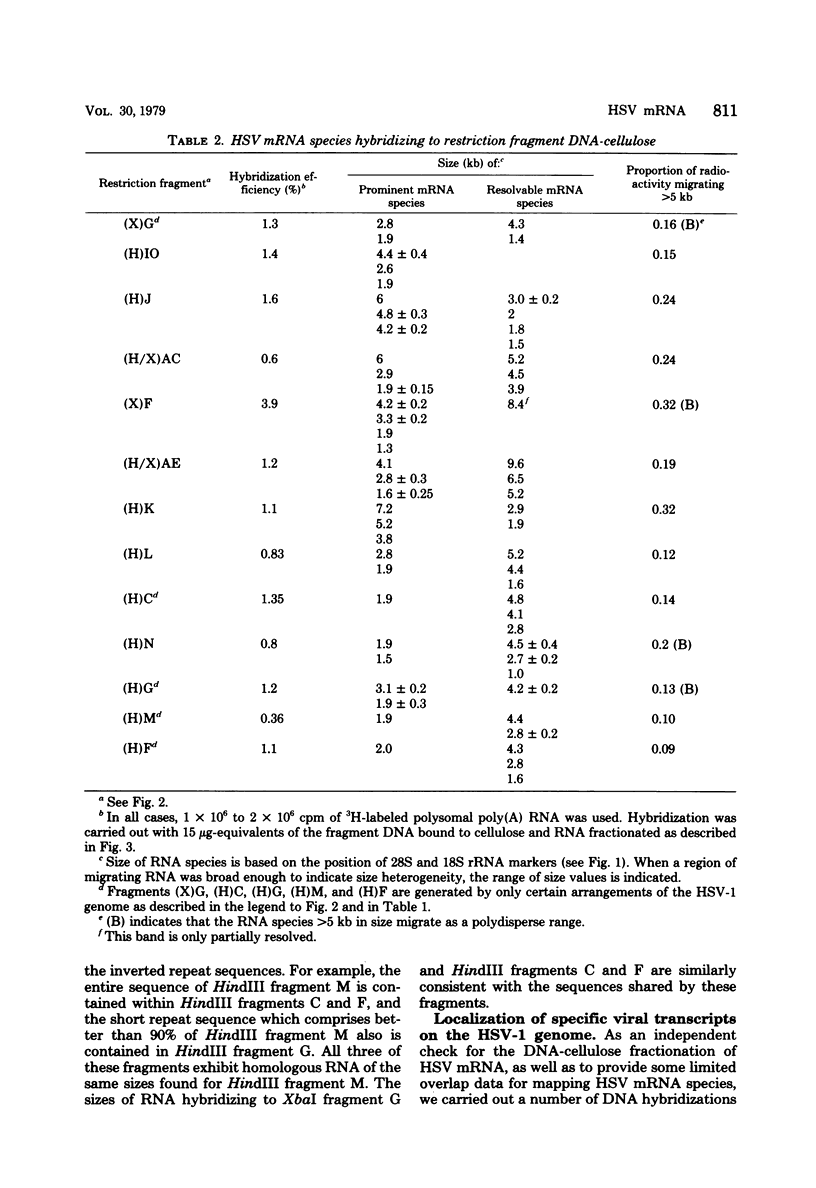
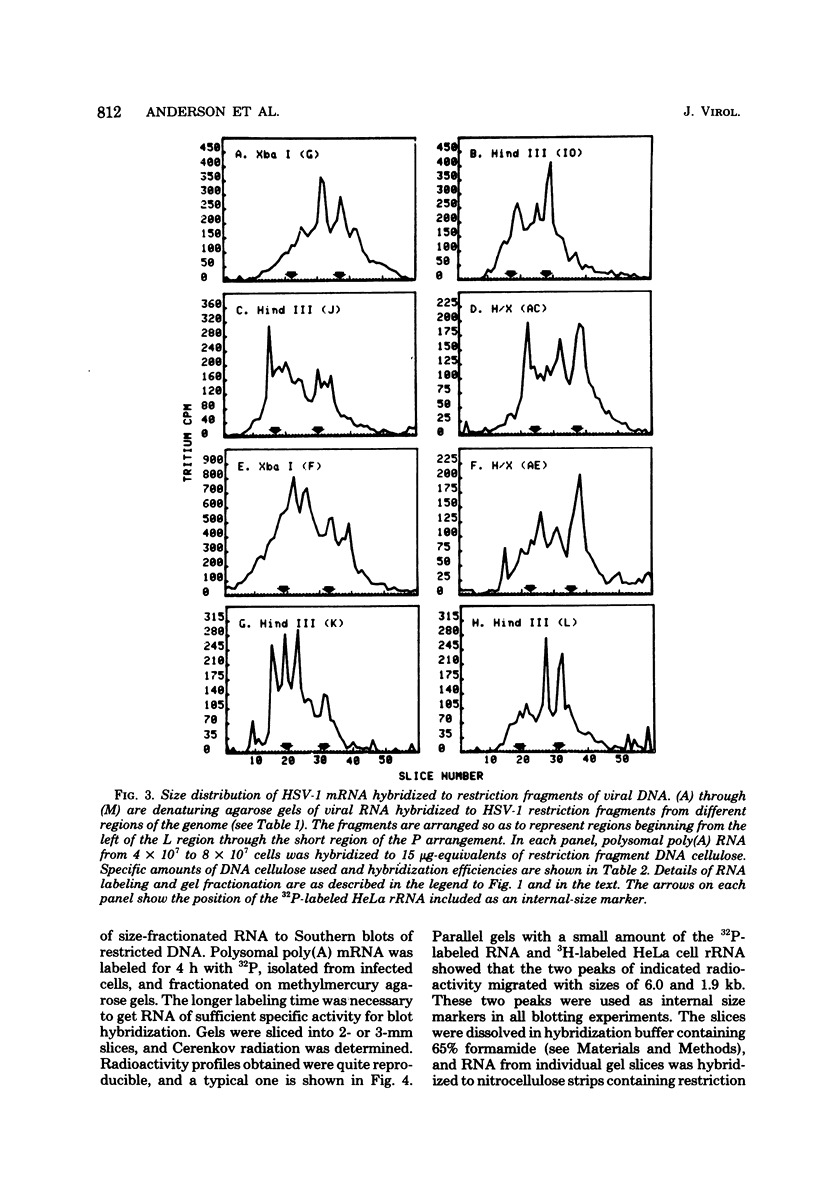
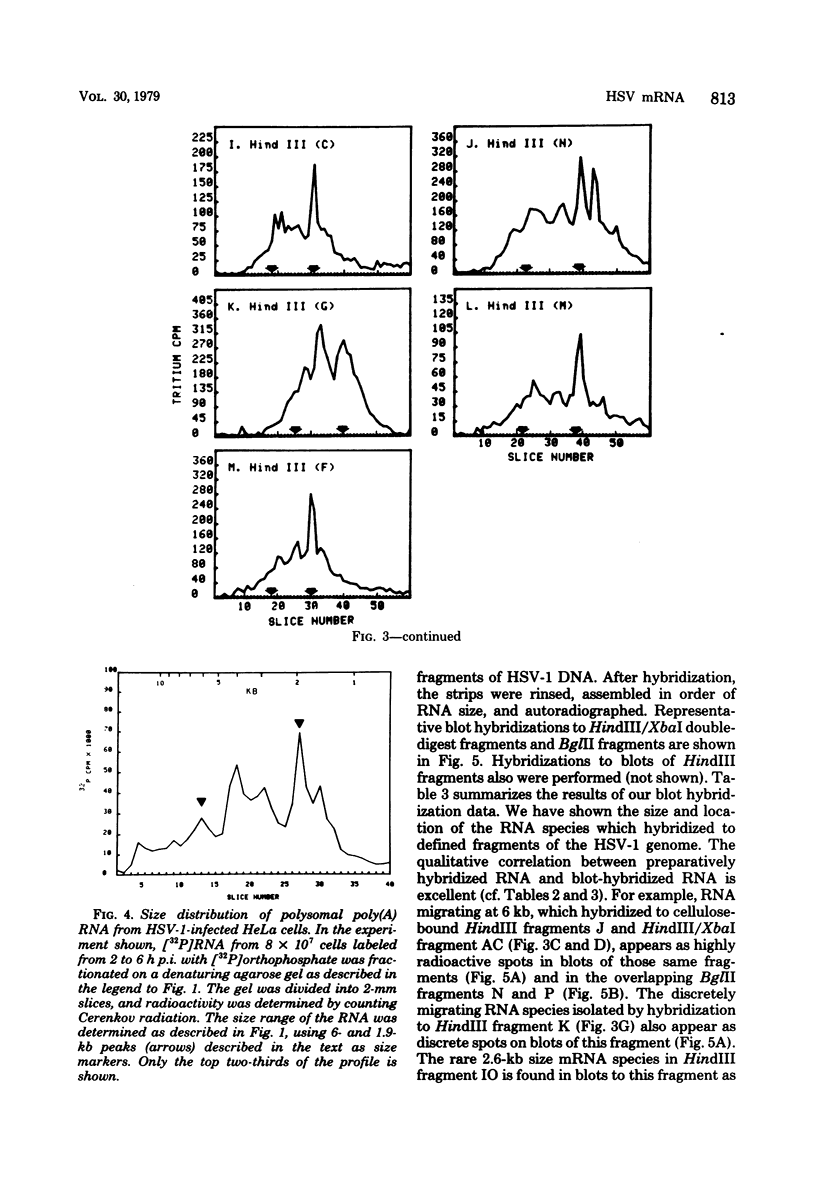
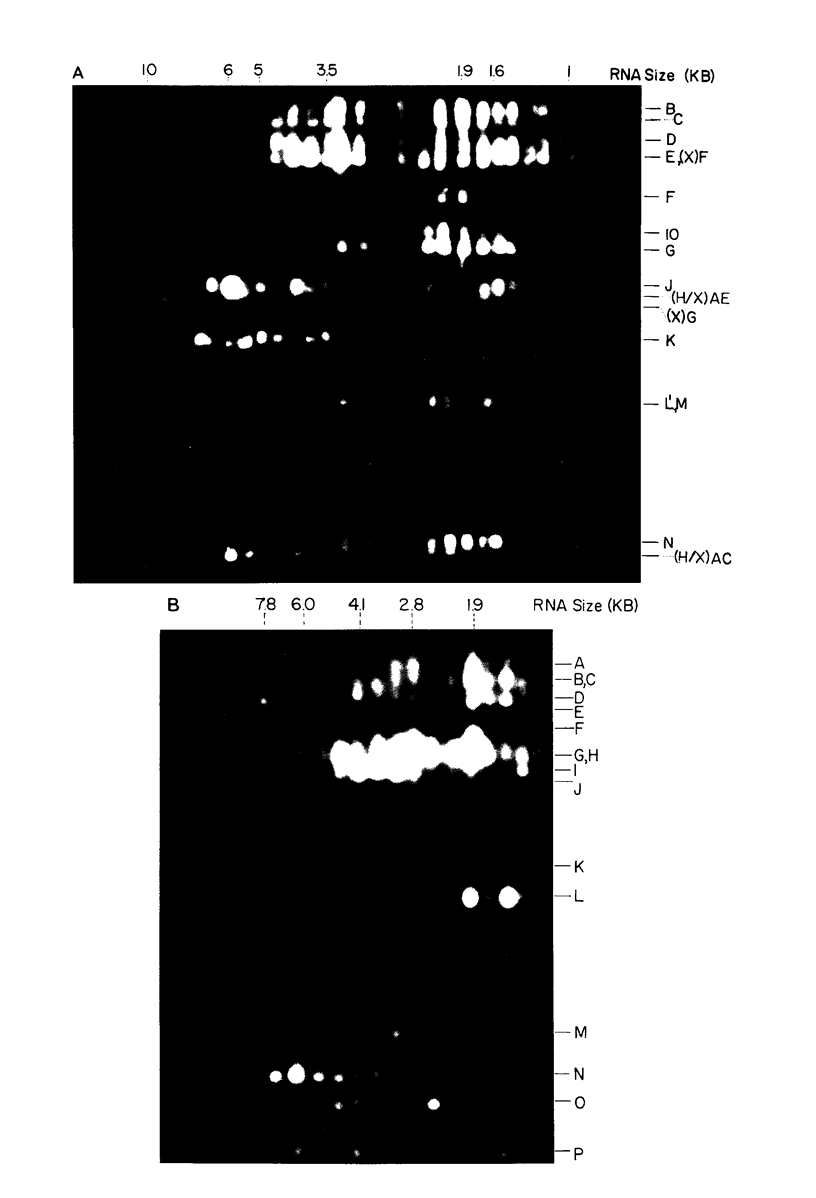
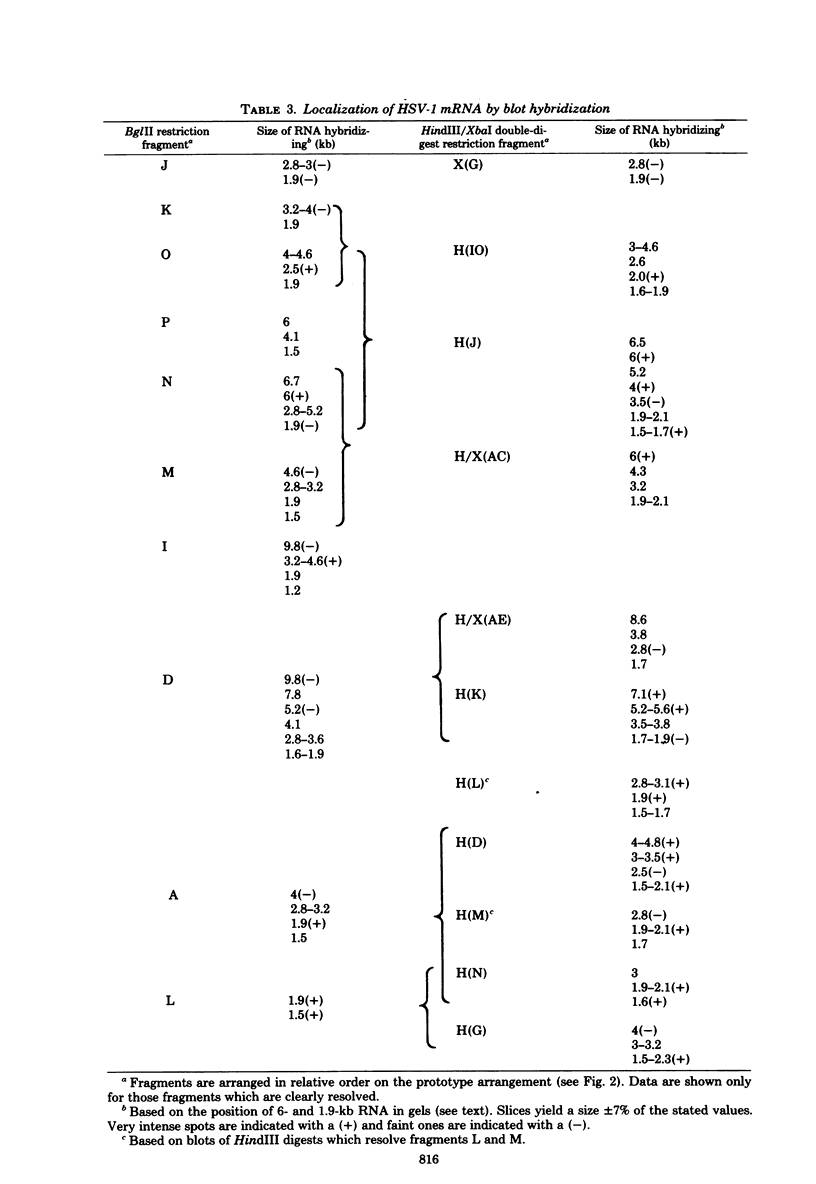
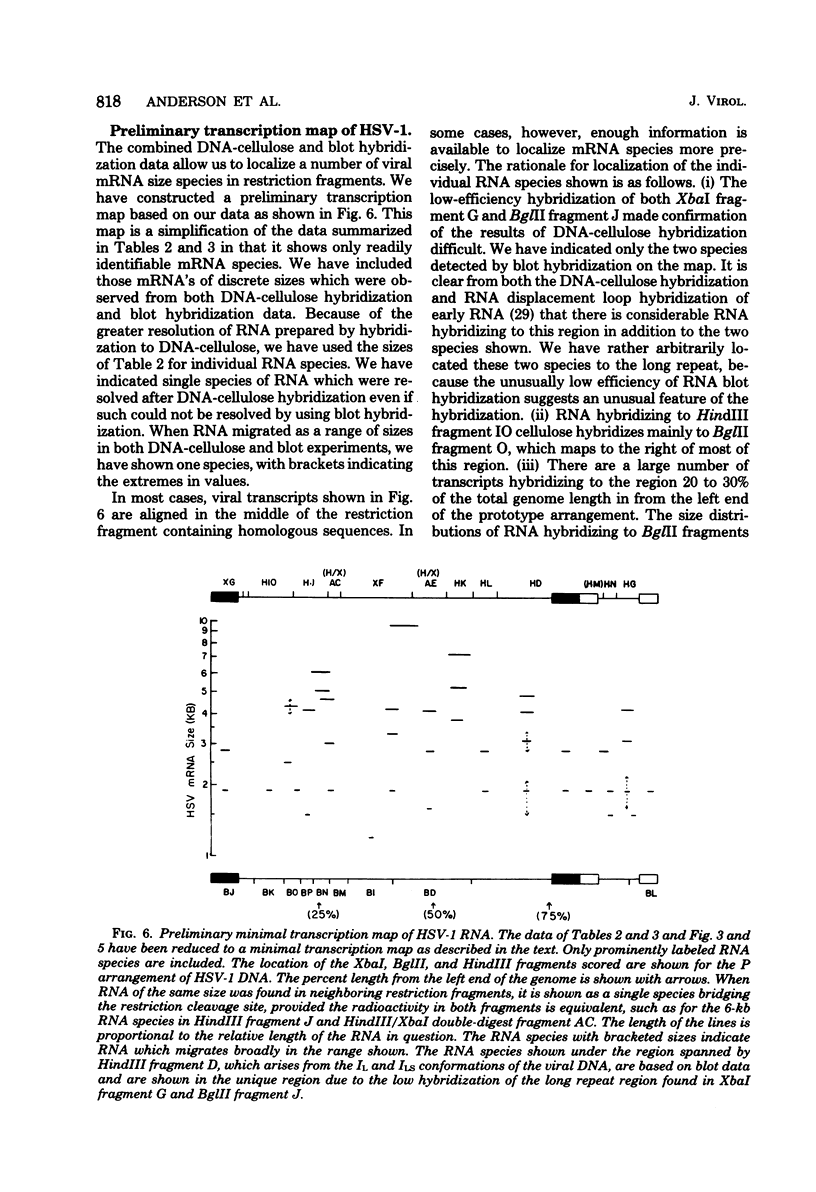
Herpes simplex virus (HSV) DNA bound to cellulose has been used as a reagent to isolate viral mRNA for size analysis on denaturing agarose gels. Total viral polysomal polyadenylated RNA was isolated from cells late after infection when such RNA has sequences encoded by approximately 45% of the HSV DNA. This RNA has a size range of from 1.5 to greater than or equal to 8 kilobases, with certain sizes, such as 1.7 to 1.9 kilobases, being favored. We have used the restriction endonucleases HindIII and XbaI singly and together to generate various sized fragments covering the entire HSV-1 genome. These fragments have been bound to cellulose to allow isolation of HSV-1 mRNA annealing to different regions of the viral genome. Discrete sizes of viral mRNA are associated with certain regions of the genome, but the mRNA population hybridizing to even the smallest restriction fragments is complex. We used hybridization of size-fractionated RNA to Southern blots of restriction fragments of HSV-1 DNA generated by the BglII as well as HindIII and XbaI endonucleases to confirm the preparative hybridization data and to provide some overlap data for positioning transcripts. The data of blot and preparative hybridization agreed very well and were combined to construct a preliminary transcription map of HSV-1. Such a map revealed at least two areas of the long unique region of the HSV-1 genome which annealed to a large number of HSV-1 transcripts. Furthermore, discrete-sized mRNA species larger than 5 kilobases in length were found only in the middle of the long unique region. The implications of these data are discussed.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aloni Y., Dhar R., Laub O., Horowitz M., Khoury G. Novel mechanism for RNA maturation: the leader sequences of simian virus 40 mRNA are not transcribed adjacent to the coding sequences. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3686–3690. doi: 10.1073/pnas.74.9.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J. M., Davidson N. Methylmercury as a reversible denaturing agent for agarose gel electrophoresis. Anal Biochem. 1976 Jan;70(1):75–85. doi: 10.1016/s0003-2697(76)80049-8. [DOI] [PubMed] [Google Scholar]
- Bartkoski M., Roizman B. RNA synthesis in cells infected with herpes simple virus. XIII. Differences in the methylation patterns of viral RNA during the reproductive cycle. J Virol. 1976 Dec;20(3):583–588. doi: 10.1128/jvi.20.3.583-588.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berget S. M., Moore C., Sharp P. A. Spliced segments at the 5' terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3171–3175. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow L. T., Gelinas R. E., Broker T. R., Roberts R. J. An amazing sequence arrangement at the 5' ends of adenovirus 2 messenger RNA. Cell. 1977 Sep;12(1):1–8. doi: 10.1016/0092-8674(77)90180-5. [DOI] [PubMed] [Google Scholar]
- Clements J. B., Watson R. J., Wilkie N. M. Temporal regulation of herpes simplex virus type 1 transcription: location of transcripts on the viral genome. Cell. 1977 Sep;12(1):275–285. doi: 10.1016/0092-8674(77)90205-7. [DOI] [PubMed] [Google Scholar]
- Grafstrom R. H., Alwine J. C., Steinhart W. L., Hill C. W., Hyman R. W. The terminal repetition of herpes simplex virus DNA. Virology. 1975 Sep;67(1):144–157. doi: 10.1016/0042-6822(75)90412-2. [DOI] [PubMed] [Google Scholar]
- Hayward G. S., Jacob R. J., Wadsworth S. C., Roizman B. Anatomy of herpes simplex virus DNA: evidence for four populations of molecules that differ in the relative orientations of their long and short components. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4243–4247. doi: 10.1073/pnas.72.11.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm C. A., Oliver S. G., Newman A. M., Holland L. E., McLaughlin C. S., Wagner E. K., Warner R. C. The molecular weight of yeast P1 double-stranded RNA. J Biol Chem. 1978 Nov 25;253(22):8332–8336. [PubMed] [Google Scholar]
- Honess R. W., Roizman B. Proteins specified by herpes simplex virus. XI. Identification and relative molar rates of synthesis of structural and nonstructural herpes virus polypeptides in the infected cell. J Virol. 1973 Dec;12(6):1347–1365. doi: 10.1128/jvi.12.6.1347-1365.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honess R. W., Roizman B. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974 Jul;14(1):8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemont B., Roizman B. RNA synthesis in cells infected with herpes simplex virus. X. Properties of viral symmetric transcripts and of double-stranded RNA prepared from them. J Virol. 1975 Apr;15(4):707–713. doi: 10.1128/jvi.15.4.707-713.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. C., Hayward G. S., Roizman B. Anatomy of herpes simplex virus DNA VII. alpha-RNA is homologous to noncontiguous sites in both the L and S components of viral DNA. J Virol. 1977 Jan;21(1):268–276. doi: 10.1128/jvi.21.1.268-276.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M., Roizman B. Regulation of herpesvirus macromolecular synthesis: nuclear retention of nontranslated viral RNA sequences. Proc Natl Acad Sci U S A. 1974 Nov;71(11):4322–4326. doi: 10.1073/pnas.71.11.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C. J., Dhar R., Khoury G. Mapping the spliced and unspliced late lytic SV40 RNAs. Cell. 1978 Aug;14(4):971–982. doi: 10.1016/0092-8674(78)90351-3. [DOI] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse L. S., Buchman T. G., Roizman B., Schaffer P. A. Anatomy of herpes simplex virus DNA. IX. Apparent exclusion of some parental DNA arrangements in the generation of intertypic (HSV-1 X HSV-2) recombinants. J Virol. 1977 Oct;24(1):231–248. doi: 10.1128/jvi.24.1.231-248.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse L. S., Pereira L., Roizman B., Schaffer P. A. Anatomy of herpes simplex virus (HSV) DNA. X. Mapping of viral genes by analysis of polypeptides and functions specified by HSV-1 X HSV-2 recombinants. J Virol. 1978 May;26(2):389–410. doi: 10.1128/jvi.26.2.389-410.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B., Gershowitz A., Stringer J. R., Holland L. E., Wagner E. K. 5'-Terminal and internal methylated nucleosides in herpes simplex virus type 1 mRNA. J Virol. 1977 Aug;23(2):234–239. doi: 10.1128/jvi.23.2.234-239.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes B. E., Stark G. R. Nucleic acid hybridization using DNA covalently coupled to cellulose. Cell. 1975 Jul;5(3):301–310. doi: 10.1016/0092-8674(75)90105-1. [DOI] [PubMed] [Google Scholar]
- Palmiter R. D. Magnesium precipitation of ribonucleoprotein complexes. Expedient techniques for the isolation of undergraded polysomes and messenger ribonucleic acid. Biochemistry. 1974 Aug 13;13(17):3606–3615. doi: 10.1021/bi00714a032. [DOI] [PubMed] [Google Scholar]
- Plummer G., Goodheart C. R., Henson D., Bowling C. P. A comparative study of the DNA density and behavior in tissue cultures of fourteen different herpesviruses. Virology. 1969 Sep;39(1):134–137. doi: 10.1016/0042-6822(69)90355-9. [DOI] [PubMed] [Google Scholar]
- Sheldrick P., Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Silvertien S., Millette R., Jones P., Roizman B. RNA synthesis in cells infected with herpes simplex virus. XII. Sequence complexity and properties of RNA differing in extent of adenylation. J Virol. 1976 Jun;18(3):977–991. doi: 10.1128/jvi.18.3.977-991.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skare J., Summers W. C. Structure and function of herpesvirus genomes. II. EcoRl, Sbal, and HindIII endonuclease cleavage sites on herpes simplex virus. Virology. 1977 Feb;76(2):581–595. doi: 10.1016/0042-6822(77)90240-9. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stringer J. R., Holland L. E., Swanstrom R. I., Pivo K., Wagner E. K. Quantitation of herpes simplex virus type 1 RNA in infected HeLa cells. J Virol. 1977 Mar;21(3):889–901. doi: 10.1128/jvi.21.3.889-901.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer J. R., Holland L. E., Wagner E. K. Mapping early transcripts of herpes simplex virus type 1 by electron microscopy. J Virol. 1978 Jul;27(1):56–73. doi: 10.1128/jvi.27.1.56-73.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanstrom R. I., Pivo K., Wagner E. K. Restricted transcription of the herpes simplex virus genome occurring early after infection and in the presence of metabolic inhibitors. Virology. 1975 Jul;66(1):140–150. doi: 10.1016/0042-6822(75)90185-3. [DOI] [PubMed] [Google Scholar]
- Swanstrom R. I., Wagner E. K. Regulation of synthesis of herpes simplex type 1 virus mRNA during productive infection. Virology. 1974 Aug;60(2):522–533. doi: 10.1016/0042-6822(74)90346-8. [DOI] [PubMed] [Google Scholar]
- Wadsworth S., Jacob R. J., Roizman B. Anatomy of herpes simplex virus DNA. II. Size, composition, and arrangement of inverted terminal repetitions. J Virol. 1975 Jun;15(6):1487–1497. doi: 10.1128/jvi.15.6.1487-1497.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E. K., Swanstrom R. I., Rice M., Howell L., Lane J. Variation in the molecular size of the DNA from closely related strains of type I herpes simplex virus. Biochim Biophys Acta. 1976 Jun 18;435(2):192–205. doi: 10.1016/0005-2787(76)90250-1. [DOI] [PubMed] [Google Scholar]
- Wagner E. K., Swanstrom R. I., Stafford M. G. Transcription of the herpes simplex virus genome in human cells. J Virol. 1972 Oct;10(4):675–682. doi: 10.1128/jvi.10.4.675-682.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E. K., Tewari K. K., Kolodner R., Warner R. C. The molecular size of the herpes simplex virus type 1 genome. Virology. 1974 Feb;57(2):436–447. doi: 10.1016/0042-6822(74)90183-4. [DOI] [PubMed] [Google Scholar]
- Weiss S. R., Varmus H. E., Bishop J. M. The size and genetic composition of virus-specific RNAs in the cytoplasm of cells producing avian sarcoma-leukosis viruses. Cell. 1977 Dec;12(4):983–992. doi: 10.1016/0092-8674(77)90163-5. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B. Secondary structure maps of RNA: processing of HeLa ribosomal RNA. Proc Natl Acad Sci U S A. 1973 Oct;70(10):2827–2831. doi: 10.1073/pnas.70.10.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie N. M., Cortini R. Sequence arrangement in herpes simplex virus type 1 DNA: identification of terminal fragments in restriction endonuclease digests and evidence for inversions in redundant and unique sequences. J Virol. 1976 Oct;20(1):211–221. doi: 10.1128/jvi.20.1.211-221.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie N. M. Physical maps for Herpes simplex virus type 1 DNA for restriction endonucleases Hind III, Hpa-1, and X. bad. J Virol. 1976 Oct;20(1):222–233. doi: 10.1128/jvi.20.1.222-233.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]