Abstract
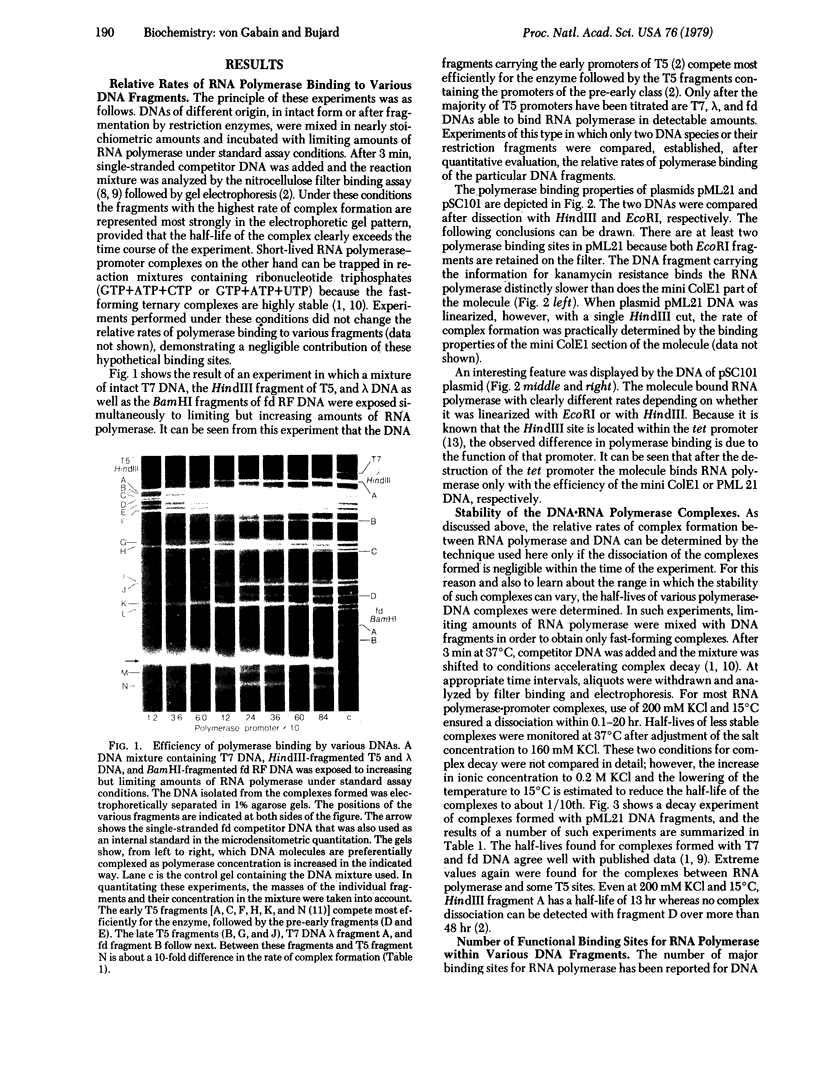
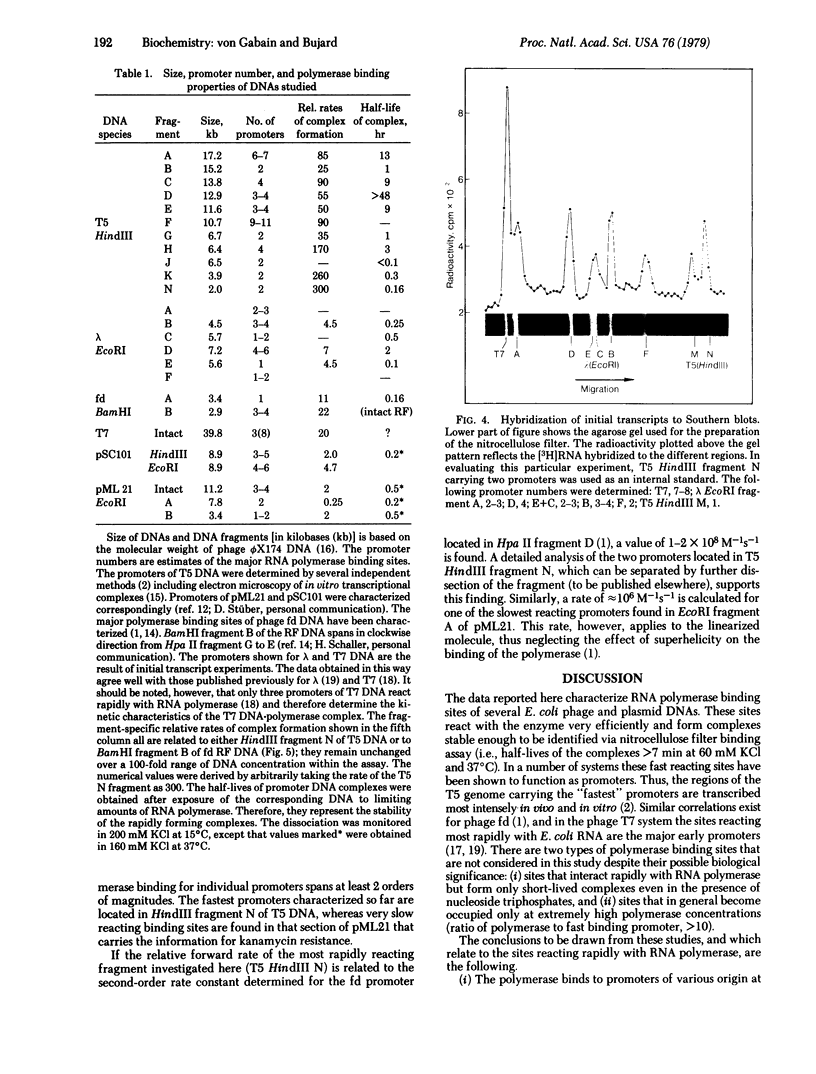
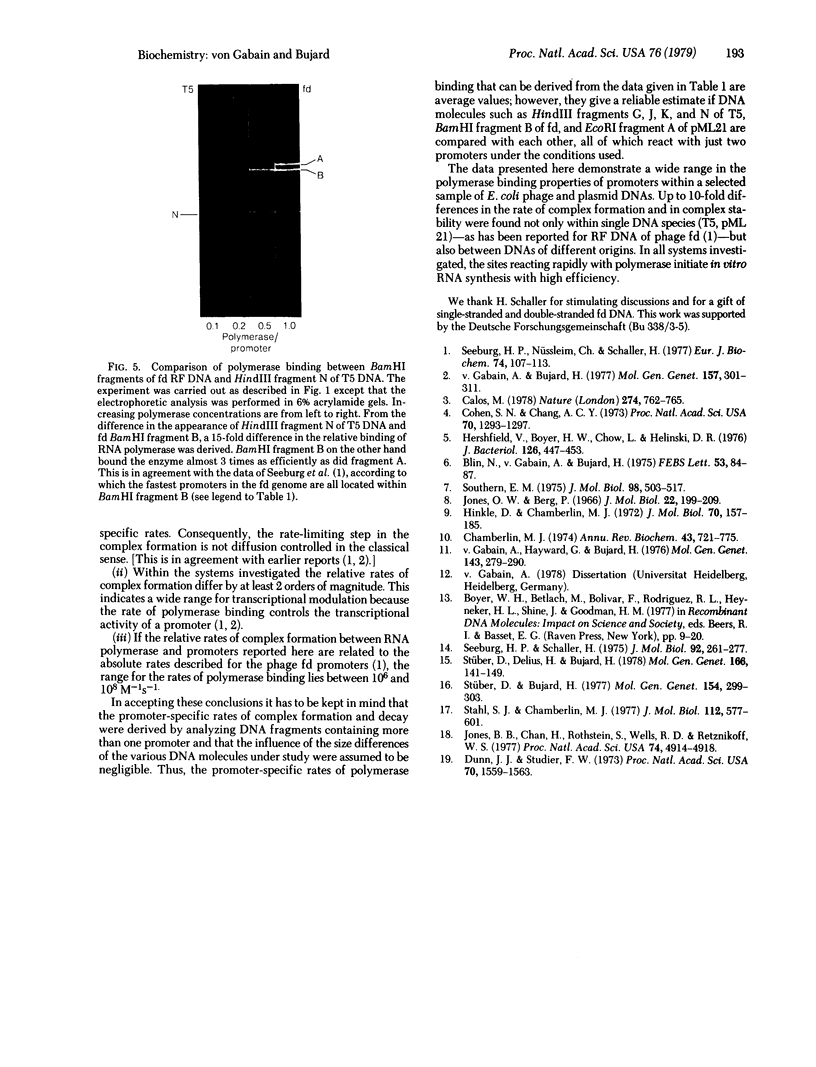
The interaction of Escherichia coli RNA polymerase (nucleosidetriphosphate:RNA nucleotidyltransferase, EC 2.7.7.6) with restriction fragments obtained from various E. coli related DNAs was studied in vitro. The DNAs investigated included several coliphage genomes (T5, lambda, T7, fd) and plasmid DNAs (pML 21, pSC101). By using the nitrocellulose filter binding of the enzyme-DNA complexes, fragment-specific relative rates of complex formation as well as complex stabilities were determined. Promoter-specific relative rates of polymerase binding were derived from fragment-specific rates by taking into account the number of major binding sites for RNA polymerase within several DNAs. Estimates of the stability of complexes formed between some major binding sites and the enzyme were obtained by studying the rate of complex decay. Both characteristics--rate of complex formation and rate of decay--varied widely and independently of each other. The promoters reacting most efficiently with E. coli RNA polymerase were found in the early region of coliphage T5 whereas some promoters in pML 21, or for example, the lambda promoter PI, belong to signals binding the enzyme most slowly. Based on the second-order rate constant determined for the interaction of E. coli RNA polymerase with promoters of phage fd, the fastest promoters characterized so far reacted with rates in the order of 10(8) M-1s-1. The hierarchy of promoters established here is of interest from the viewpoint that promoter strength correlates with the rate of polymerase binding. Among the promoters studied here this rate spans a range of 2 orders of magnitude.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Calos M. P. DNA sequence for a low-level promoter of the lac repressor gene and an 'up' promoter mutation. Nature. 1978 Aug 24;274(5673):762–765. doi: 10.1038/274762a0. [DOI] [PubMed] [Google Scholar]
- Chamberlin M. J. The selectivity of transcription. Annu Rev Biochem. 1974;43(0):721–775. doi: 10.1146/annurev.bi.43.070174.003445. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C. Recircularization and autonomous replication of a sheared R-factor DNA segment in Escherichia coli transformants. Proc Natl Acad Sci U S A. 1973 May;70(5):1293–1297. doi: 10.1073/pnas.70.5.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. T7 early RNAs are generated by site-specific cleavages. Proc Natl Acad Sci U S A. 1973 May;70(5):1559–1563. doi: 10.1073/pnas.70.5.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershfield V., Boyer H. W., Chow L., Helinski D. R. Characterization of a mini-ColC1 plasmid. J Bacteriol. 1976 Apr;126(1):447–453. doi: 10.1128/jb.126.1.447-453.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinkle D. C., Chamberlin M. J. Studies of the binding of Escherichia coli RNA polymerase to DNA. I. The role of sigma subunit in site selection. J Mol Biol. 1972 Sep 28;70(2):157–185. doi: 10.1016/0022-2836(72)90531-1. [DOI] [PubMed] [Google Scholar]
- Jones B. B., Chan H., Rothstein S., Wells R. D., Reznikoff W. S. RNA polymerase binding sites in lambdaplac5 DNA. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4914–4918. doi: 10.1073/pnas.74.11.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones O. W., Berg P. Studies on the binding of RNA polymerase to polynucleotides. J Mol Biol. 1966 Dec 28;22(2):199–209. doi: 10.1016/0022-2836(66)90126-4. [DOI] [PubMed] [Google Scholar]
- Seeburg P. H., Nüsslein C., Schaller H. Interaction of RNA polymerase with promoters from bacteriophage fd. Eur J Biochem. 1977 Mar 15;74(1):107–113. doi: 10.1111/j.1432-1033.1977.tb11372.x. [DOI] [PubMed] [Google Scholar]
- Seeburg P. H., Schaller H. Mapping and characterization of promoters in bacteriophages fd, f1 and m13. J Mol Biol. 1975 Feb 25;92(2):261–277. doi: 10.1016/0022-2836(75)90226-0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stahl S. J., Chamberlin M. J. An expanded transcriptional map of T7 bacteriophage. Reading of minor T7 promoter sites in vitro by Escherichia coli RNA polymerase. J Mol Biol. 1977 Jun 5;112(4):577–601. doi: 10.1016/s0022-2836(77)80165-4. [DOI] [PubMed] [Google Scholar]
- Stüber D., Bujard H. Electron microscopy of DNA: determination of absolute molecular weights and linear density. Mol Gen Genet. 1977 Sep 9;154(3):299–303. doi: 10.1007/BF00571286. [DOI] [PubMed] [Google Scholar]
- Stüber D., Delius H., Bujard H. Electron microscopic analysis of in vitro transcriptional complexes: mapping of promoters of the coliphage T5 genome. Mol Gen Genet. 1978 Oct 30;166(2):141–149. doi: 10.1007/BF00285916. [DOI] [PubMed] [Google Scholar]
- von Gabain A., Bujard H. Interaction of E. coli RNA polymerase with promotors of coliphage T5: the rates of complex formation and decay and their correlation with in vitro and in vivo transcriptional activity. Mol Gen Genet. 1977 Dec 9;157(3):301–311. doi: 10.1007/BF00268667. [DOI] [PubMed] [Google Scholar]
- von Gabain A., Hayward G. S., Bujard H. Physical mapping of the HindIII, EcoRI, Sal and Sma restriction endonuclease cleavage fragments from bacteriophage T5 DNA. Mol Gen Genet. 1976 Feb 2;143(3):279–290. doi: 10.1007/BF00269404. [DOI] [PubMed] [Google Scholar]