Abstract
Rat-liver chromatin has bee fractionated into transcriptionally active and inactive regions [Gottesfeld et al. (1974) Proc. Nat. Acad. Sci. USA 71, 2193-2197] and the distribution of nuclease-resistant complexes in these fractions has been investigated. About half of the DNA of both fractions is resistant to attack by tne endonuclease DNase II. The nuclease-resistant structures of inactive chromatin are DNA-histone complexes (v-bodies) which sediment at 11-13 S. Template-active chromatin yields two peaks of nuclease-resistant nucleoprotein. These complexes sediment at 14 and 19 S, and contain DNA, RNA, histone, and nonhistone chromosomal proteins. Polyacrylamide gel electrophoresis reveals a complex pattern of chromatin proteins, suggesting that the complexes are heterogeneous in composition.
Full text
PDF

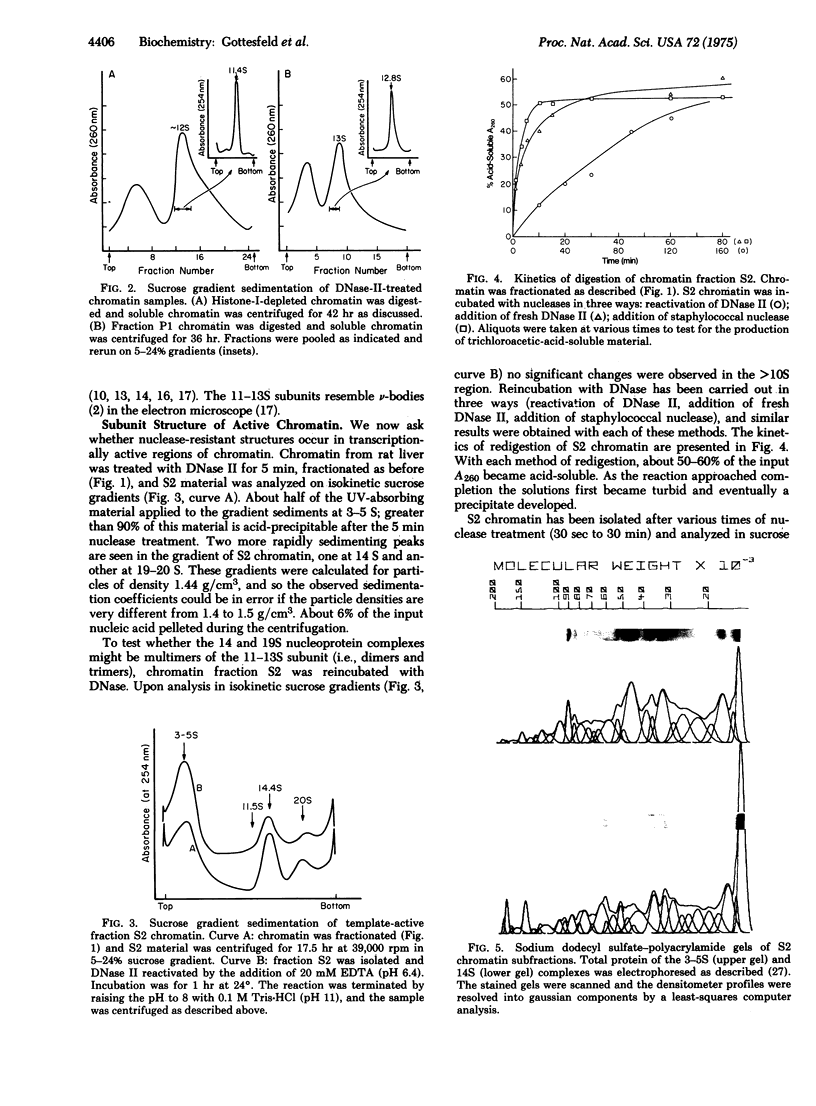


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Axel R., Cedar H., Felsenfeld G. Chromatin template activity and chromatin structure. Cold Spring Harb Symp Quant Biol. 1974;38:773–783. doi: 10.1101/sqb.1974.038.01.082. [DOI] [PubMed] [Google Scholar]
- Axel R., Melchior W., Jr, Sollner-Webb B., Felsenfeld G. Specific sites of interaction between histones and DNA in chromatin. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4101–4105. doi: 10.1073/pnas.71.10.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billing R. J., Bonner J. The structure of chromatin as revealed by deoxyribonuclease digestion studies. Biochim Biophys Acta. 1972 Oct 27;281(3):453–462. doi: 10.1016/0005-2787(72)90462-5. [DOI] [PubMed] [Google Scholar]
- Burgoyne L. A., Hewish D. R., Mobbs J. Mammalian chromatin substructure studies with the calcium-magnesium endonuclease and two-dimensional polyacrylamide-gel electrophoresis. Biochem J. 1974 Oct;143(1):67–72. doi: 10.1042/bj1430067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R. J., Felsenfeld G. Chemical probes of chromatin structure. Biochemistry. 1974 Aug 13;13(17):3622–3628. doi: 10.1021/bi00714a034. [DOI] [PubMed] [Google Scholar]
- Clark R. J., Felsenfeld G. Structure of chromatin. Nat New Biol. 1971 Jan 27;229(4):101–106. doi: 10.1038/newbio229101a0. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J. M., Bonner J., Radda G. K., Walker I. O. Biophysical studies on the mechanism of quinacrine staining of chromosomes. Biochemistry. 1974 Jul 2;13(14):2937–2945. doi: 10.1021/bi00711a600. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J. M., Garrard W. T., Bagi G., Wilson R. F., Bonner J. Partial purification of the template-active fraction of chromatin: a preliminary report. Proc Natl Acad Sci U S A. 1974 Jun;71(6):2193–2197. doi: 10.1073/pnas.71.6.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J. D. Chromatin structure: deduced from a minichromosome. Science. 1975 Mar 28;187(4182):1202–1203. doi: 10.1126/science.187.4182.1202. [DOI] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- King J., Laemmli U. K. Polypeptides of the tail fibres of bacteriophage T4. J Mol Biol. 1971 Dec 28;62(3):465–477. doi: 10.1016/0022-2836(71)90148-3. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Chromatin structure: a repeating unit of histones and DNA. Science. 1974 May 24;184(4139):868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Marushige K., Bonner J. Fractionation of liver chromatin. Proc Natl Acad Sci U S A. 1971 Dec;68(12):2941–2944. doi: 10.1073/pnas.68.12.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marushige K., Bonner J. Template properties of liver chromatin. J Mol Biol. 1966 Jan;15(1):160–174. doi: 10.1016/s0022-2836(66)80218-8. [DOI] [PubMed] [Google Scholar]
- McConaughy B. L., McCarthy B. J. Fractionation of chromatin by thermal chromatography. Biochemistry. 1972 Mar 14;11(6):998–1003. doi: 10.1021/bi00756a008. [DOI] [PubMed] [Google Scholar]
- Miller O. L., Jr Fine structure of lampbrush chromosomes. Natl Cancer Inst Monogr. 1965 Dec;18:79–99. [PubMed] [Google Scholar]
- Noll H. Characterization of macromolecules by constant velocity sedimentation. Nature. 1967 Jul 22;215(5099):360–363. doi: 10.1038/215360a0. [DOI] [PubMed] [Google Scholar]
- Noll M. Subunit structure of chromatin. Nature. 1974 Sep 20;251(5472):249–251. doi: 10.1038/251249a0. [DOI] [PubMed] [Google Scholar]
- Olins A. L., Olins D. E. Spheroid chromatin units (v bodies). Science. 1974 Jan 25;183(4122):330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- Oosterhof D. K., Hozier J. C., Rill R. L. Nucleas action on chromatin: evidence for discrete, repeated nucleoprotein units along chromatin fibrils. Proc Natl Acad Sci U S A. 1975 Feb;72(2):633–637. doi: 10.1073/pnas.72.2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudet P., Gross-Bellard M., Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975 Apr;4(4):281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- Panyim S., Chalkley R. High resolution acrylamide gel electrophoresis of histones. Arch Biochem Biophys. 1969 Mar;130(1):337–346. doi: 10.1016/0003-9861(69)90042-3. [DOI] [PubMed] [Google Scholar]
- Pardon J. F., Wilkins M. H., Richards B. M. Super-helical model for nucleohistone. Nature. 1967 Jul 29;215(5100):508–509. doi: 10.1038/215508a0. [DOI] [PubMed] [Google Scholar]
- Polacow I., Simpson R. T. Circular dichroism spectra of putative transcribed and repressed chromatin. Biochem Biophys Res Commun. 1973 May 1;52(1):202–207. doi: 10.1016/0006-291x(73)90974-1. [DOI] [PubMed] [Google Scholar]
- Rill R., Van Holde K. E. Properties of nuclease-resistant fragments of calf thymus chromatin. J Biol Chem. 1973 Feb 10;248(3):1080–1083. [PubMed] [Google Scholar]
- STUDIER F. W. SEDIMENTATION STUDIES OF THE SIZE AND SHAPE OF DNA. J Mol Biol. 1965 Feb;11:373–390. doi: 10.1016/s0022-2836(65)80064-x. [DOI] [PubMed] [Google Scholar]
- Sahasrabuddhe C. G., Van Holde K. E. The effect of trypsin on nuclease-resistant chromatin fragments. J Biol Chem. 1974 Jan 10;249(1):152–156. [PubMed] [Google Scholar]
- Senior M. B., Olins A. L., Olins D. E. Chromatin fragments resembling v bodies. Science. 1975 Jan 17;187(4172):173–175. doi: 10.1126/science.1111096. [DOI] [PubMed] [Google Scholar]
- van Bruggen E. F., Arnberg A. C., van Holde K. E., Sahasrabuddhe C. G., Shaw B. R. Electron microscopy of chromatin subunit particles. Biochem Biophys Res Commun. 1974 Oct 23;60(4):1365–1370. doi: 10.1016/0006-291x(74)90348-9. [DOI] [PubMed] [Google Scholar]