Abstract
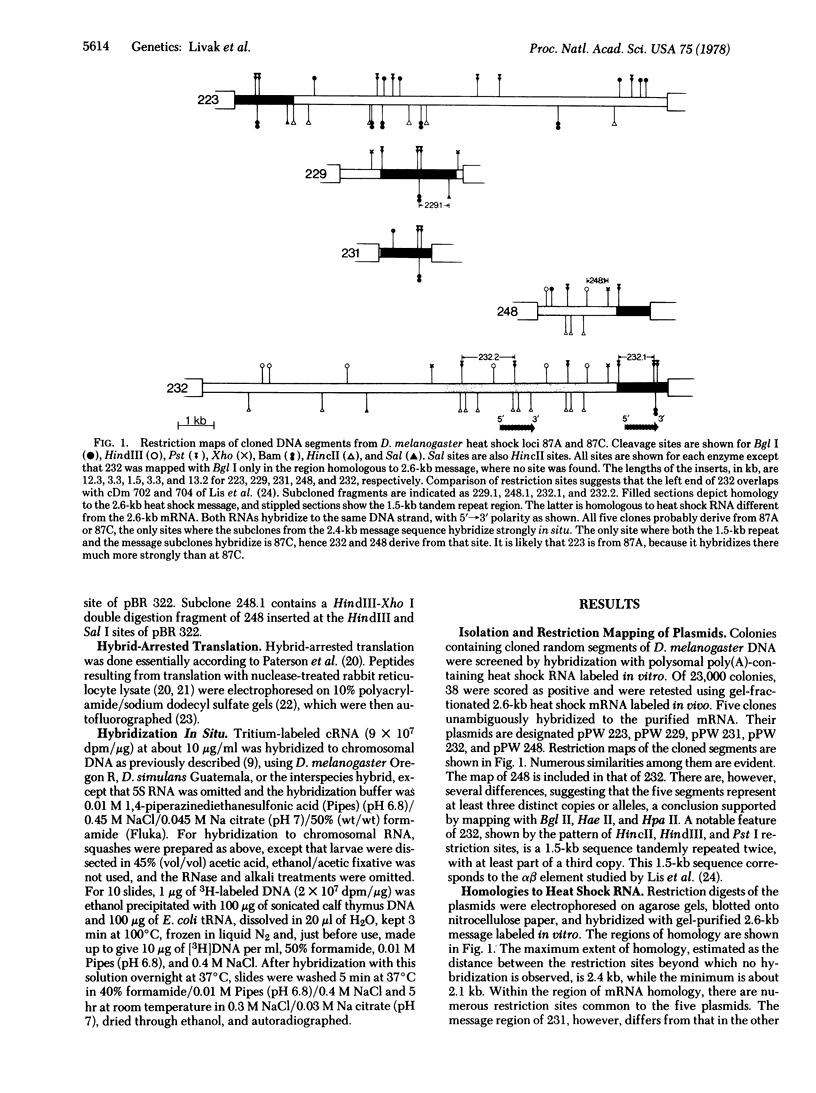
The heat shock loci of Drosophila melanogaster chromosome subdivisions 87A and 87C have been studied by using cloned DNA. Both sites contain a number of copies of a 2,4-kilobase (kb) region homologous to mRNA for the 70,000-dalton heat shock protein. In situ hybridization to chromosomal RNA shows that transcripts of this sequence accumulate at both sites after temperature elevation. At 87C there is a 1.5-kb repeated sequence homologous to another heat shock RNA. One cloned segment includes two to three tandem copies of this sequence located 0.8 kb from the beginning of a 2.4-kb message region. RNA complementary to the 1.5-kb repeat acccumulates at 87C after temperature elevation, but does not code for any known heat shock protein. In the sibling species D. simulans, there are sequences located and transcribed at 87A and 87C that are homologous to the melanogaster 2.4-kb message sequence. The 1.5-kb repeat, however, is absent from 87C in simulans and no heat shock RNA homologous to it can be detected.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ashburner M. Patterns of puffing activity in the salivary gland chromosomes of Drosophila. V. Responses to environmental treatments. Chromosoma. 1970;31(3):356–376. doi: 10.1007/BF00321231. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Botchan M., Topp W., Sambrook J. The arrangement of simian virus 40 sequences in the DNA of transformed cells. Cell. 1976 Oct;9(2):269–287. doi: 10.1016/0092-8674(76)90118-5. [DOI] [PubMed] [Google Scholar]
- Clewell D. B. Nature of Col E 1 plasmid replication in Escherichia coli in the presence of the chloramphenicol. J Bacteriol. 1972 May;110(2):667–676. doi: 10.1128/jb.110.2.667-676.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Meselson M. Transcription at two heat shock loci in Drosophila. Cell. 1977 Oct;12(2):441–451. doi: 10.1016/0092-8674(77)90120-9. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Holden J. J., Gehring W. J. Deletions of two heat-activated loci in Drosophila melanogaster and their effects on heat-induced protein synthesis. Cell. 1977 Nov;12(3):643–652. doi: 10.1016/0092-8674(77)90264-1. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lis J. T., Prestidge L., Hogness D. S. A novel arrangement of tandemly repeated genes at a major heat shock site in D. melanogaster. Cell. 1978 Aug;14(4):901–919. doi: 10.1016/0092-8674(78)90345-8. [DOI] [PubMed] [Google Scholar]
- Lubsen N. H., Sondermeijer P. J., Pages M., Alonso C. In situ hybridization of nuclear and cytoplasmic RNA to locus 2-48BC in Drosophila hydei. Chromosoma. 1978 Feb 10;65(3):199–212. doi: 10.1007/BF00327617. [DOI] [PubMed] [Google Scholar]
- Maizels N. Dictyostelium 17S, 25S, and 5S rDNAs lie within a 38,000 base pair repeated unit. Cell. 1976 Nov;9(3):431–438. doi: 10.1016/0092-8674(76)90088-x. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- McKenzie S. L., Henikoff S., Meselson M. Localization of RNA from heat-induced polysomes at puff sites in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1117–1121. doi: 10.1073/pnas.72.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie S. L., Meselson M. Translation in vitro of Drosophila heat-shock messages. J Mol Biol. 1977 Nov 25;117(1):279–283. doi: 10.1016/0022-2836(77)90035-3. [DOI] [PubMed] [Google Scholar]
- Mirault M. E., Goldschmidt-Clermont M., Moran L., Arrigo A. P., Tissières A. The effect of heat shock on gene expression in Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):819–827. doi: 10.1101/sqb.1978.042.01.082. [DOI] [PubMed] [Google Scholar]
- Murray K., Murray N. E. Phage lambda receptor chromosomes for DNA fragments made with restriction endonuclease III of Haemophilus influenzae and restriction endonuclease I of Escherichia coli. J Mol Biol. 1975 Nov 5;98(3):551–564. doi: 10.1016/s0022-2836(75)80086-6. [DOI] [PubMed] [Google Scholar]
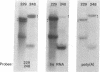
- Paterson B. M., Roberts B. E., Kuff E. L. Structural gene identification and mapping by DNA-mRNA hybrid-arrested cell-free translation. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4370–4374. doi: 10.1073/pnas.74.10.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham H. R., Jackson R. J. An efficient mRNA-dependent translation system from reticulocyte lysates. Eur J Biochem. 1976 Aug 1;67(1):247–256. doi: 10.1111/j.1432-1033.1976.tb10656.x. [DOI] [PubMed] [Google Scholar]
- Pukkila P. J. Identification of the lampbrush chromosome loops which transcribe 5S ribosomal RNA in Notophthalmus (Triturus) viridescens. Chromosoma. 1975 Nov 20;53(1):71–89. doi: 10.1007/BF00329391. [DOI] [PubMed] [Google Scholar]
- Schedl P., Artavanis-Tsakonas S., Steward R., Gehring W. J., Mirault M. E., Goldschmidt-Clermont M., Moran L., Tissières A. Two hybrid plasmids with D. melanogaster DNA sequences complementary to mRNA coding for the major heat shock protein. Cell. 1978 Aug;14(4):921–929. doi: 10.1016/0092-8674(78)90346-x. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spradling A., Pardue M. L., Penman S. Messenger RNA in heat-shocked Drosophila cells. J Mol Biol. 1977 Feb 5;109(4):559–587. doi: 10.1016/s0022-2836(77)80091-0. [DOI] [PubMed] [Google Scholar]
- Spradling A., Penman S., Pardue M. L. Analysis of drosophila mRNA by in situ hybridization: sequences transcribed in normal and heat shocked cultured cells. Cell. 1975 Apr;4(4):395–404. doi: 10.1016/0092-8674(75)90160-9. [DOI] [PubMed] [Google Scholar]
- Sugden B., De Troy B., Roberts R. J., Sambrook J. Agarose slab-gel electrophoresis equipment. Anal Biochem. 1975 Sep;68(1):36–46. doi: 10.1016/0003-2697(75)90676-4. [DOI] [PubMed] [Google Scholar]
- Tissières A., Mitchell H. K., Tracy U. M. Protein synthesis in salivary glands of Drosophila melanogaster: relation to chromosome puffs. J Mol Biol. 1974 Apr 15;84(3):389–398. doi: 10.1016/0022-2836(74)90447-1. [DOI] [PubMed] [Google Scholar]
- Wensink P. C., Finnegan D. J., Donelson J. E., Hogness D. S. A system for mapping DNA sequences in the chromosomes of Drosophila melanogaster. Cell. 1974 Dec;3(4):315–325. doi: 10.1016/0092-8674(74)90045-2. [DOI] [PubMed] [Google Scholar]