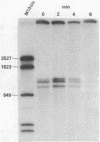
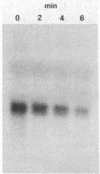
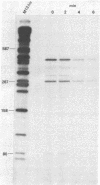
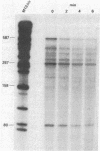
Abstract
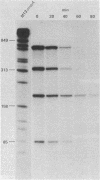
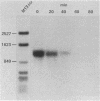
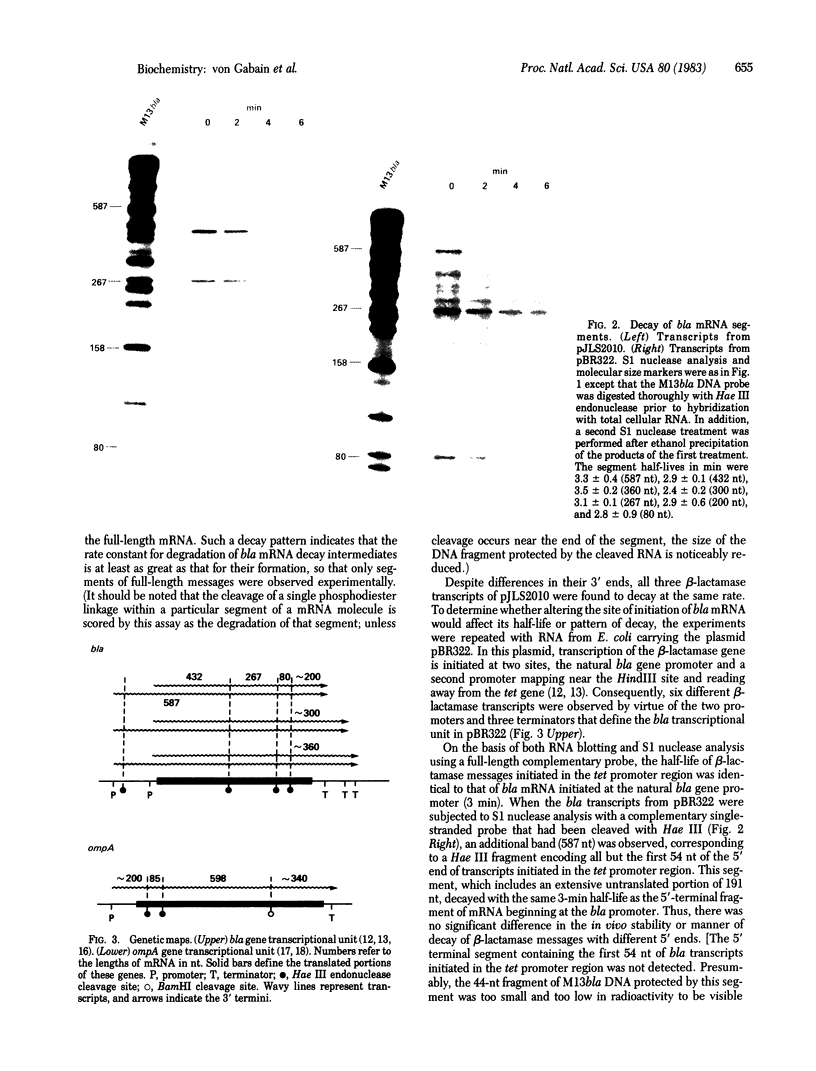
An assay was developed to investigate the fate of specific segments of beta-lactamase (bla) and ompA gene transcripts in Escherichia coli. DNA probes cloned in bacteriophage M13 were treated with an endonuclease capable of cleaving single-stranded DNA, the fragments produced were annealed with total cellular RNA, and the resulting RNA . DNA hybrids were subjected to S1 nuclease treatment and gel fractionation. By using this assay, direct evidence was obtained for 3'-to-5' directionality in the decay of the long-lived mRNA encoded by the ompA gene, and no preferential stability was observed for translated versus untranslated mRNA segments. In the case of bla mRNA, initial cleavage of the full-length transcript was rate limiting, and no decay intermediates were detected. No difference in degradation rate was seen for bla transcripts having variant 3' or 5' termini.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beck E., Bremer E. Nucleotide sequence of the gene ompA coding the outer membrane protein II of Escherichia coli K-12. Nucleic Acids Res. 1980 Jul 11;8(13):3011–3027. doi: 10.1093/nar/8.13.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Blakesley R. W., Dodgson J. B., Nes I. F., Wells R. D. Duplex regions in "single-stranded" phiX174 DNA are cleaved by a restriction endonuclease from Haemophilus aegyptius. J Biol Chem. 1977 Oct 25;252(20):7300–7306. [PubMed] [Google Scholar]
- Blundell M., Kennell D. Evidence for endonucleolytic attack in decay of lac messenger RNA in Escherichia coli. J Mol Biol. 1974 Feb 25;83(2):143–161. doi: 10.1016/0022-2836(74)90385-4. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Bremer E., Beck E., Hindennach I., Sonntag I., Henning U. Cloned structural gene (ompA) for an integral outer membrane protein of Escherichia coli K-12: localization on hybrid plasmid pTU100 and expression of a fragment of the gene. Mol Gen Genet. 1980;179(1):13–20. doi: 10.1007/BF00268440. [DOI] [PubMed] [Google Scholar]
- Brosius J., Cate R. L., Perlmutter A. P. Precise location of two promoters for the beta-lactamase gene of pBR322. S1 mapping of ribonucleic acid isolated from Escherichia coli or synthesized in vitro. J Biol Chem. 1982 Aug 10;257(15):9205–9210. [PubMed] [Google Scholar]
- Chang A. C., Erlich H. A., Gunsalus R. P., Nunberg J. H., Kaufman R. J., Schimke R. T., Cohen S. N. Initiation of protein synthesis in bacteria at a translational start codon of mamalian cDNA: effects of the preceding nucleotide sequence. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1442–1446. doi: 10.1073/pnas.77.3.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarneros G., Montañez C., Hernandez T., Court D. Posttranscriptional control of bacteriophage lambda gene expression from a site distal to the gene. Proc Natl Acad Sci U S A. 1982 Jan;79(2):238–242. doi: 10.1073/pnas.79.2.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaguni J., Ray D. S. Cloning of a functional replication origin of phage G4 into the genome of phage M13. J Mol Biol. 1979 Dec 25;135(4):863–878. doi: 10.1016/0022-2836(79)90516-3. [DOI] [PubMed] [Google Scholar]
- Manor H., Goodman D., Stent G. S. RNA chain growth rates in Escherichia coli. J Mol Biol. 1969 Jan 14;39(1):1–29. doi: 10.1016/0022-2836(69)90329-5. [DOI] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy P. L., Symons R. H. Cleavage of DNA.RNA hybrids by type II restriction enzymes. Nucleic Acids Res. 1980 Jul 11;8(13):2939–2946. doi: 10.1093/nar/8.13.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa N., Imamoto F. Degradation of tryptophan messenger. On the degradation of messenger RNA for the tryptophan operon in Escherichia coli. Nature. 1969 Jul 5;223(5201):37–40. doi: 10.1038/223037a0. [DOI] [PubMed] [Google Scholar]
- Morse D. E., Mosteller R., Baker R. F., Yanofsky C. Direction of in vivo degradation of tryptophan messenger RNA--a correction. Nature. 1969 Jul 5;223(5201):40–43. doi: 10.1038/223040a0. [DOI] [PubMed] [Google Scholar]
- Morse D. E., Yanofsky C. Polarity and the degradation of mRNA. Nature. 1969 Oct 25;224(5217):329–331. doi: 10.1038/224329a0. [DOI] [PubMed] [Google Scholar]
- Movva N. R., Nakamura K., Inouye M. Regulatory region of the gene for the ompA protein, a major outer membrane protein of Escherichia coli. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3845–3849. doi: 10.1073/pnas.77.7.3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen S., Reeh S. Functional mRNA half lives in E. coli. Mol Gen Genet. 1978 Nov 9;166(3):329–336. doi: 10.1007/BF00267626. [DOI] [PubMed] [Google Scholar]
- Rubens C., Heffron F., Falkow S. Transposition of a plasmid deoxyribonucleic acid sequence that mediates ampicillin resistance: independence from host rec functions and orientation of insertion. J Bacteriol. 1976 Oct;128(1):425–434. doi: 10.1128/jb.128.1.425-434.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHERRER K., DARNELL J. E. Sedimentation characteristics of rapidly labelled RNA from HeLa cells. Biochem Biophys Res Commun. 1962 Jun 4;7:486–490. doi: 10.1016/0006-291x(62)90341-8. [DOI] [PubMed] [Google Scholar]
- Seed B. Diazotizable arylamine cellulose papers for the coupling and hybridization of nucleic acids. Nucleic Acids Res. 1982 Mar 11;10(5):1799–1810. doi: 10.1093/nar/10.5.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stüber D., Bujard H. Organization of transcriptional signals in plasmids pBR322 and pACYC184. Proc Natl Acad Sci U S A. 1981 Jan;78(1):167–171. doi: 10.1073/pnas.78.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- von Gabain A., Bujard H. Interaction of Escherichia coli RNA polymerase with promoters of several coliphage and plasmid DNAs. Proc Natl Acad Sci U S A. 1979 Jan;76(1):189–193. doi: 10.1073/pnas.76.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]