Abstract
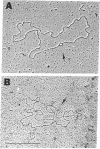
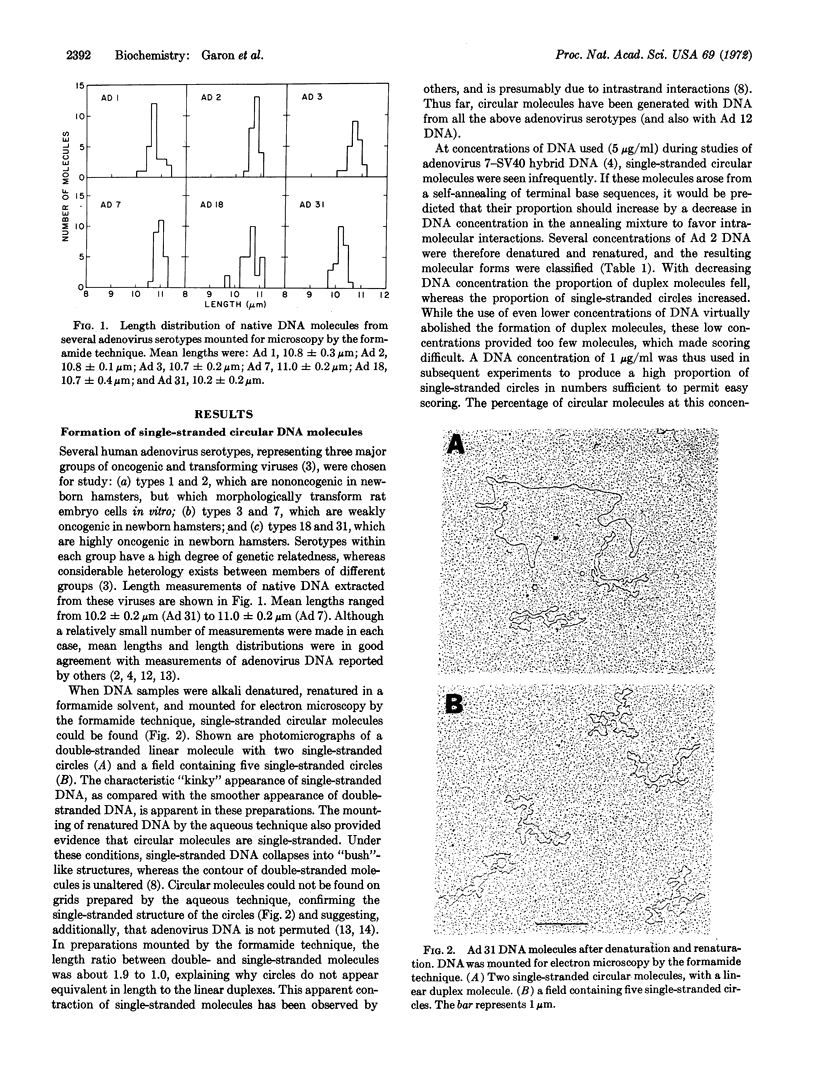
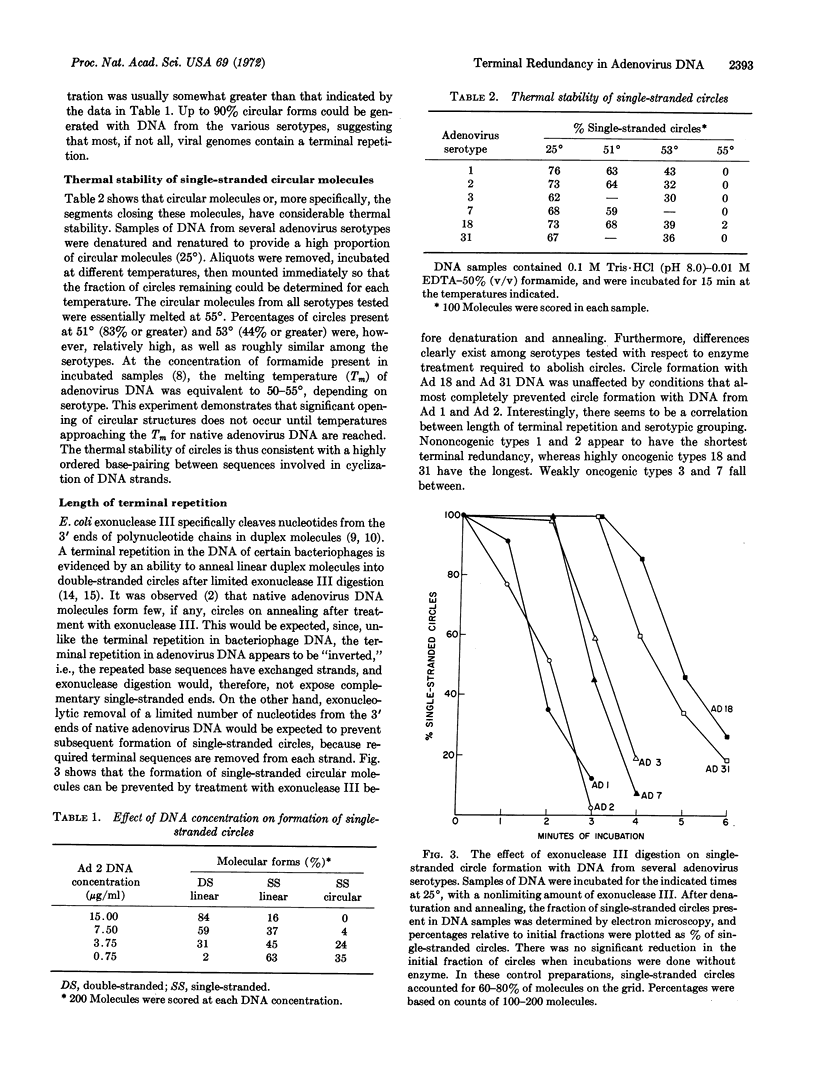
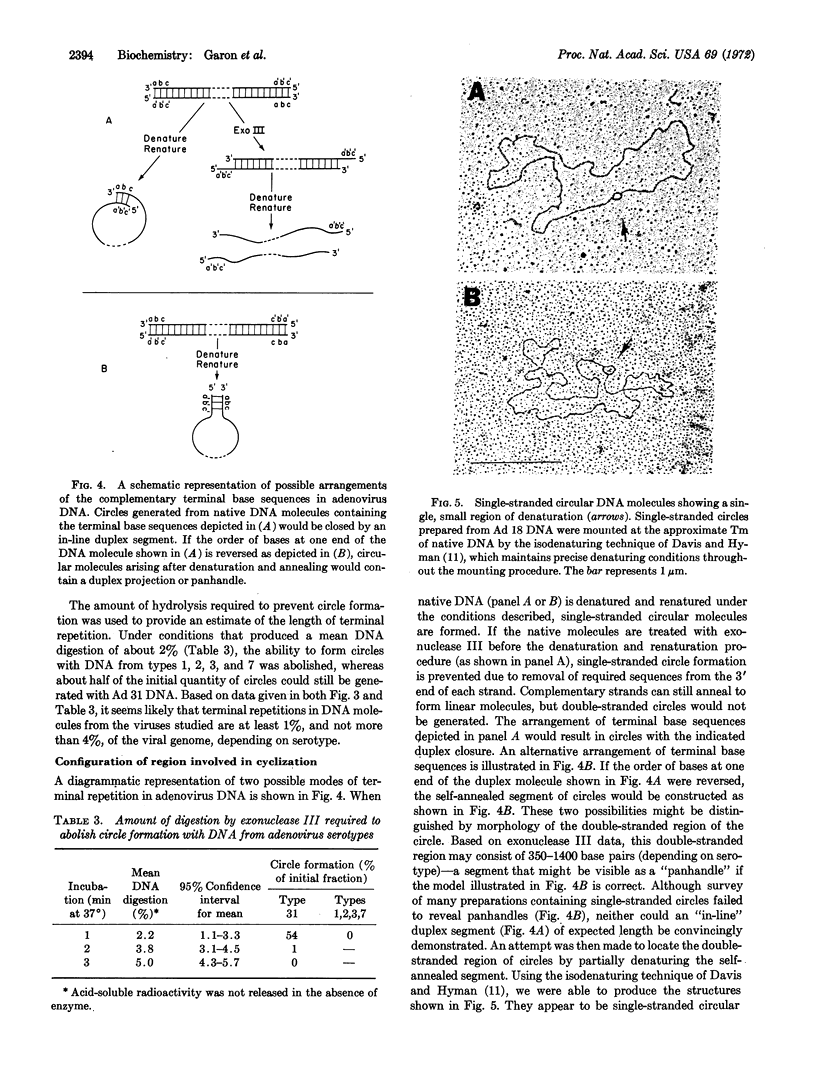
A unique form of terminal redundancy has been observed in DNA molecules extracted from several human adenovirus serotypes. Electron microscopic studies reveal that single-stranded circular molecules are formed when native DNA is denatured and then annealed. Temperatures approaching the Tm of native DNA are required to convert circles to linear molecules, indicating a high degree of self-complementarity between terminal base sequences of DNA strands. Single-stranded circles are not generated if a limited number of nucleotides (2-4%) are removed from the 3′ ends of native DNA by digestion with Escherichia coli exonuclease III before denaturation and annealing. The lenght of the redundant segment appears to differ among major serotypic groups, and a possible association between increased length of the redundant segment and increased oncogenic capability of virus serotype is suggested. Evidence for the configuration of the duplex closure region of circular molecules is also presented.
Keywords: circular molecules, self-annealing, exonuclease, electron microscopy
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Coulian M. Initiation of the replication of single-stranded DNA by Escherichia coli DNA polymerase. Cold Spring Harb Symp Quant Biol. 1968;33:11–20. doi: 10.1101/sqb.1968.033.01.006. [DOI] [PubMed] [Google Scholar]
- Davis R. W., Hyman R. W. A study in evolution: the DNA base sequence homology between coliphages T7 and T3. J Mol Biol. 1971 Dec 14;62(2):287–301. doi: 10.1016/0022-2836(71)90428-1. [DOI] [PubMed] [Google Scholar]
- Doerfler W., Hellmann W., Kleinschmidt A. K. The DNA of adenovirus type 12 and its denaturation pattern. Virology. 1972 Feb;47(2):507–512. doi: 10.1016/0042-6822(72)90290-5. [DOI] [PubMed] [Google Scholar]
- Doerfler W., Kleinschmidt A. K. Denaturation pattern of the DNA of adenovirus type 2 as determined by electron microscopy. J Mol Biol. 1970 Jun 28;50(3):579–593. doi: 10.1016/0022-2836(70)90086-0. [DOI] [PubMed] [Google Scholar]
- Green M., Piña M., Kimes R., Wensink P. C., MacHattie L. A., Thomas C. A., Jr Adenovirus DNA. I. Molecular weight and conformation. Proc Natl Acad Sci U S A. 1967 May;57(5):1302–1309. doi: 10.1073/pnas.57.5.1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly T. J., Jr, Rose J. A. Simian virus 40 integration site in an adenovirus 7-simian virus 40 hybrid DNA molecule. Proc Natl Acad Sci U S A. 1971 May;68(5):1037–1041. doi: 10.1073/pnas.68.5.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacHattie L. A., Ritchie D. A., Thomas C. A., Jr, Richardson C. C. Terminal repetition in permuted T2 bacteriophage DNA molecules. J Mol Biol. 1967 Feb 14;23(3):355–363. doi: 10.1016/s0022-2836(67)80110-4. [DOI] [PubMed] [Google Scholar]
- RICHARDSON C. C., KORNBERG A. A DEOXYRIBONUCLEIC ACID PHOSPHATASE-EXONUCLEASE FROM ESCHERICHIA COLI. I. PURIFICATION OF THE ENZYME AND CHARACTERIZATION OF THE PHOSPHATASE ACTIVITY. J Biol Chem. 1964 Jan;239:242–250. [PubMed] [Google Scholar]
- RICHARDSON C. C., LEHMAN I. R., KORNBERG A. A DEOXYRIBONUCLEIC ACID PHOSPHATASE-EXONUCLEASE FROM ESCHERICHIA COLI. II. CHARACTERIZATION OF THE EXONUCLEASE ACTIVITY. J Biol Chem. 1964 Jan;239:251–258. [PubMed] [Google Scholar]
- Ritchie D. A., Thomas C. A., Jr, MacHattie L. A., Wensink P. C. Terminal repetition in non-permuted T3 and T7 bacteriophage DNA molecules. J Mol Biol. 1967 Feb 14;23(3):365–376. doi: 10.1016/s0022-2836(67)80111-6. [DOI] [PubMed] [Google Scholar]
- Rose J. A., Berns K. I., Hoggan M. D., Koczot F. J. Evidence for a single-stranded adenovirus-associated virus genome: formation of a DNA density hybrid on release of viral DNA. Proc Natl Acad Sci U S A. 1969 Nov;64(3):863–869. doi: 10.1073/pnas.64.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose J. A., Hoggan M. D., Shatkin A. J. Nucleic acid from an adeno-associated virus: chemical and physical studies. Proc Natl Acad Sci U S A. 1966 Jul;56(1):86–92. doi: 10.1073/pnas.56.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose J. A., Koczot F. Adenovirus-associated virus multiplication. VI. Base compostion of the deoxyribonucleic acid strand species and strand-specific in vivo transcription. J Virol. 1971 Nov;8(5):771–777. doi: 10.1128/jvi.8.5.771-777.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Eb A. J., van Kesteren L. W., van Bruggen E. F. Structural properties of adenovirus DNA's. Biochim Biophys Acta. 1969 Jun 17;182(2):530–541. doi: 10.1016/0005-2787(69)90205-6. [DOI] [PubMed] [Google Scholar]