Abstract
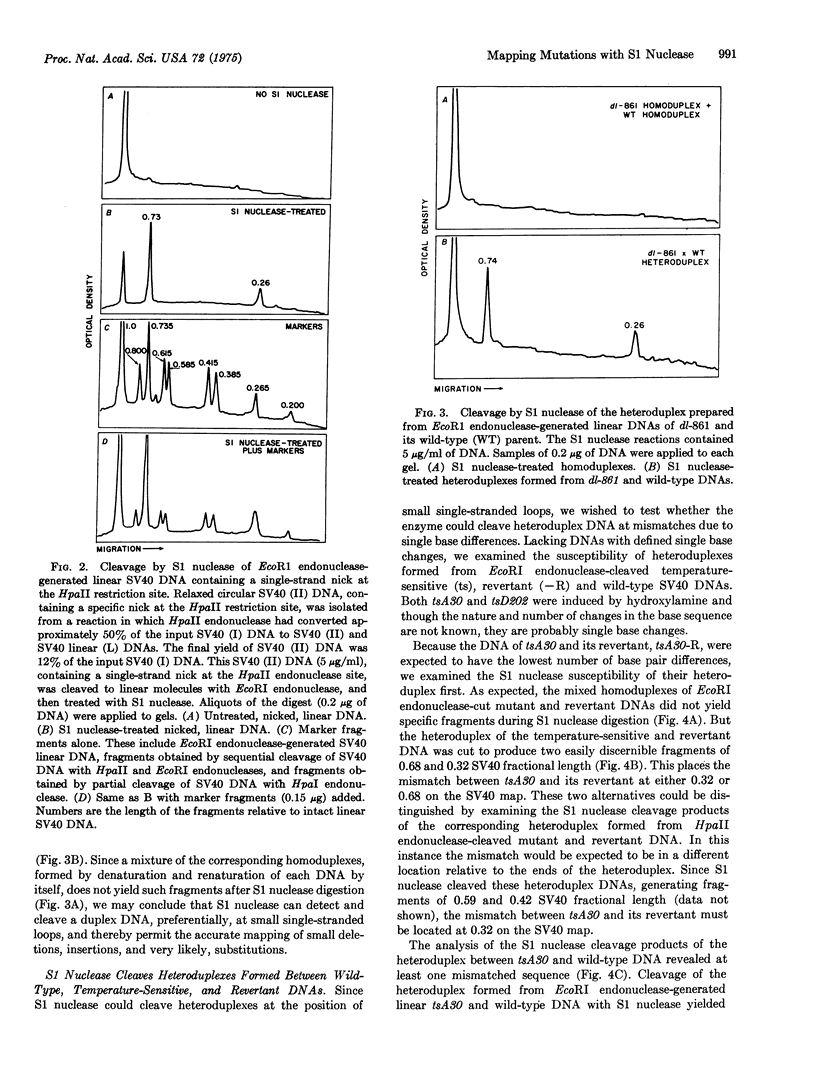
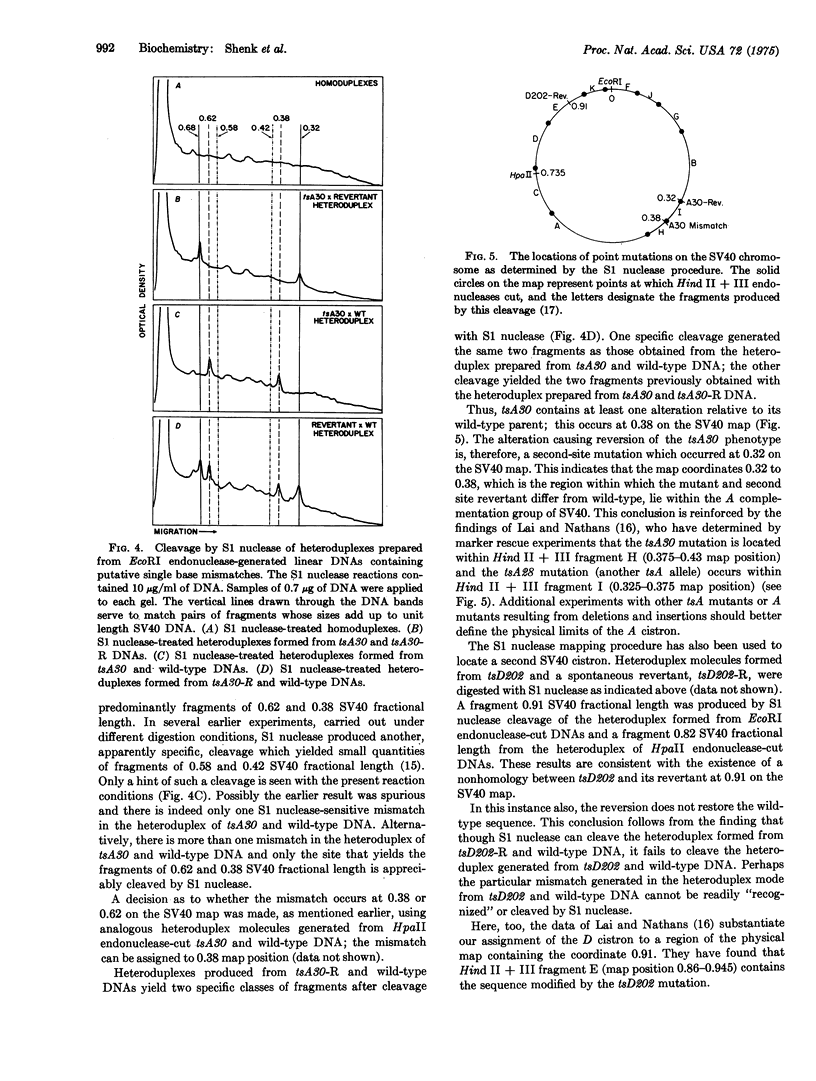
S1 nuclease (EC 3.1.4.X), a single-strand-specific nuclease, can be used to accurately map the location of mutational alterations in simian virus 40 (SV40) DNA. Deletions of between 32 and 190 base pairs, which are at or below the limit of detectability by conventional electron microscopic analysis of heteroduplex DNAs, have been located in this way. To map a deletion, a mixture of unit length, linear DNA, prepared from the SV40 deletion mutant and its wild-type parent, are denatured and reannealed to form heteroduplexes. S1 nuclease can cut such heteroduplexes at the nonbase-paired region to produce fragments whose lengths correspond to the position of the deletion. Similarly, specific fragments are produced when S1 nuclease cleaves a heteroduplex formed from the DNAs of SV40 temperature-sensitive mutants and either their revertants or wild-type parents. Thus, the positions of the nonhomology between these DNAs can be determined.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ando T. A nuclease specific for heat-denatured DNA in isolated from a product of Aspergillus oryzae. Biochim Biophys Acta. 1966 Jan 18;114(1):158–168. doi: 10.1016/0005-2787(66)90263-2. [DOI] [PubMed] [Google Scholar]
- Beard P., Morrow J. F., Berg P. Cleavage of circular, superhelical simian virus 40 DNA to a linear duplex by S1 nuclease. J Virol. 1973 Dec;12(6):1303–1313. doi: 10.1128/jvi.12.6.1303-1313.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J. Y., Martin R. G. Complementation analysis of simian virus 40 mutants. J Virol. 1974 May;13(5):1101–1109. doi: 10.1128/jvi.13.5.1101-1109.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danna K. J., Sack G. H., Jr, Nathans D. Studies of simian virus 40 DNA. VII. A cleavage map of the SV40 genome. J Mol Biol. 1973 Aug 5;78(2):363–376. doi: 10.1016/0022-2836(73)90122-8. [DOI] [PubMed] [Google Scholar]
- Germond J. E., Vogt V. M., Hirt B. Characterization of the single-strand-specific nuclease S1 activity on double-stranded supercoiled polyoma DNA. Eur J Biochem. 1974 Apr 16;43(3):591–600. doi: 10.1111/j.1432-1033.1974.tb03446.x. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Lai C. J., Nathans D. Mapping temperature-sensitive mutants of simian virus 40: rescue of mutants by fragments of viral DNA. Virology. 1974 Aug;60(2):466–475. doi: 10.1016/0042-6822(74)90340-7. [DOI] [PubMed] [Google Scholar]
- Mertz J. E., Berg P. Defective simian virus 40 genomes: isolation and growth of individual clones. Virology. 1974 Nov;62(1):112–124. doi: 10.1016/0042-6822(74)90307-9. [DOI] [PubMed] [Google Scholar]
- Mertz J. E., Berg P. Viable deletion mutants of simian virus 40: selective isolation by means of a restriction endonuclease from Hemophilus parainfluenzae. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4879–4883. doi: 10.1073/pnas.71.12.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz J. E., Davis R. W. Cleavage of DNA by R 1 restriction endonuclease generates cohesive ends. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3370–3374. doi: 10.1073/pnas.69.11.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radloff R., Bauer W., Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967 May;57(5):1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Shenk T. E., Rhodes C., Rigby P. W., Berg P. Mapping of mutational alterations in DNA with S1 nuclease: the location of deletions, insertions and temperature-sensitive mutations in SV40. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):61–67. doi: 10.1101/sqb.1974.039.01.011. [DOI] [PubMed] [Google Scholar]
- Tegtmeyer P. Simian virus 40 deoxyribonucleic acid synthesis: the viral replicon. J Virol. 1972 Oct;10(4):591–598. doi: 10.1128/jvi.10.4.591-598.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt V. M. Purification and further properties of single-strand-specific nuclease from Aspergillus oryzae. Eur J Biochem. 1973 Feb 15;33(1):192–200. doi: 10.1111/j.1432-1033.1973.tb02669.x. [DOI] [PubMed] [Google Scholar]


