Abstract
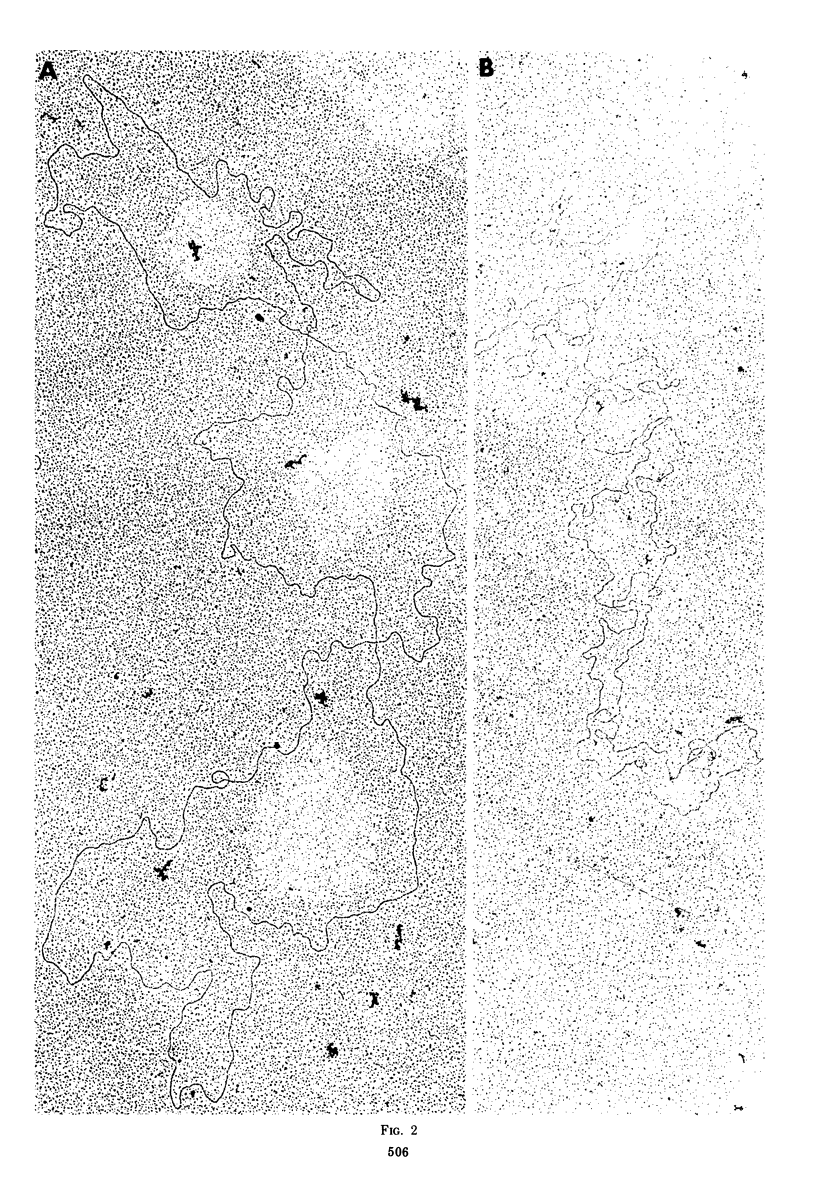
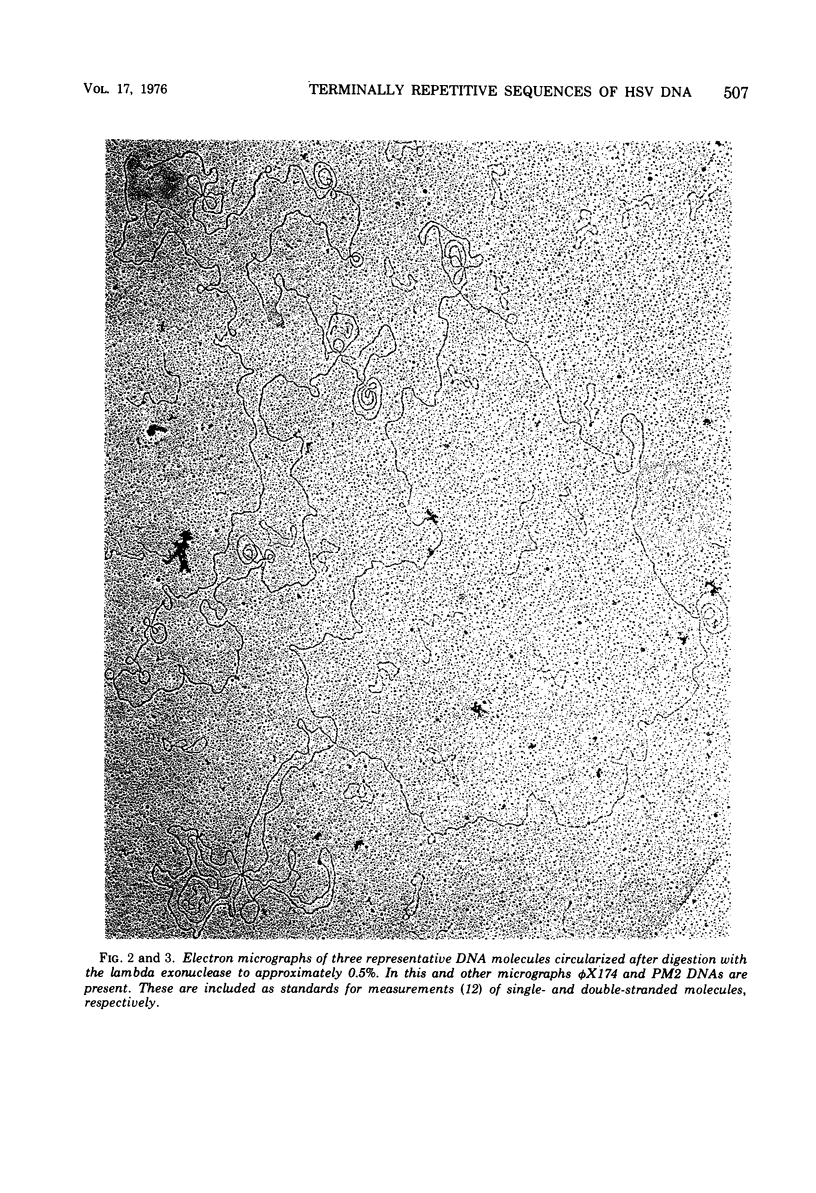
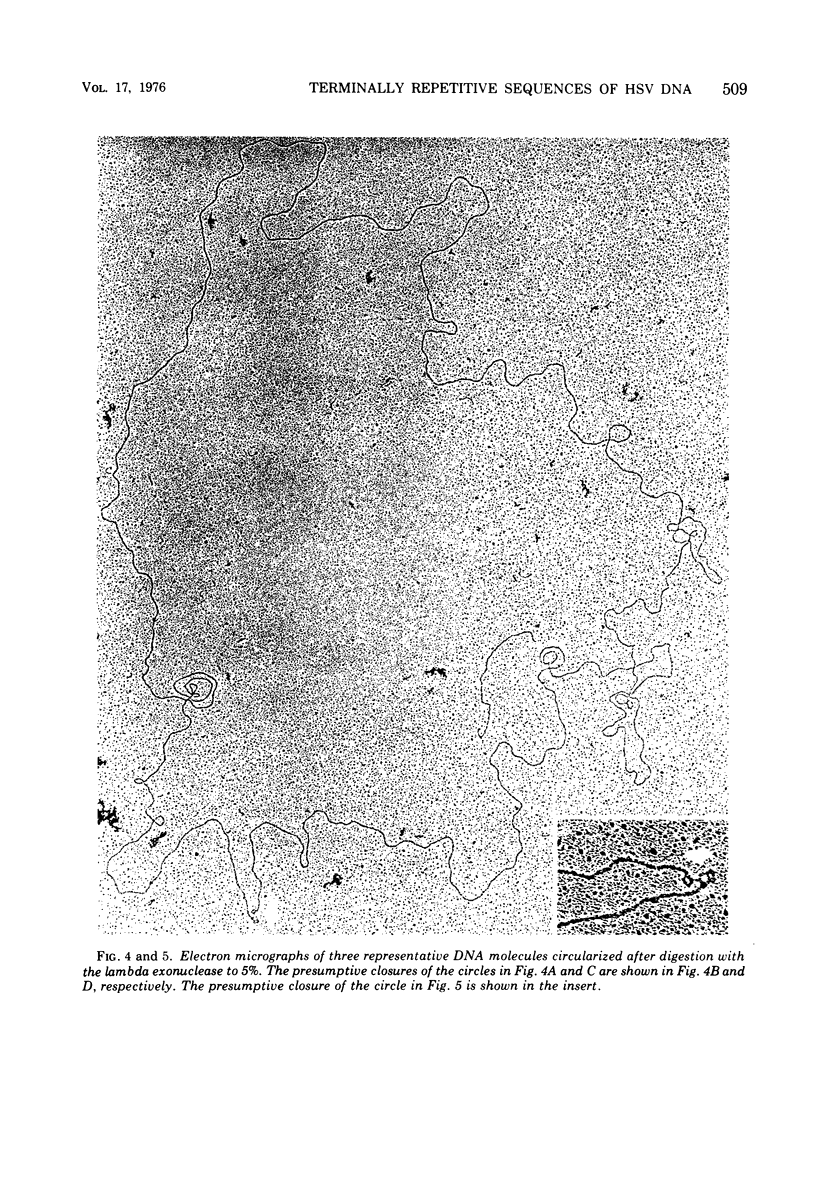
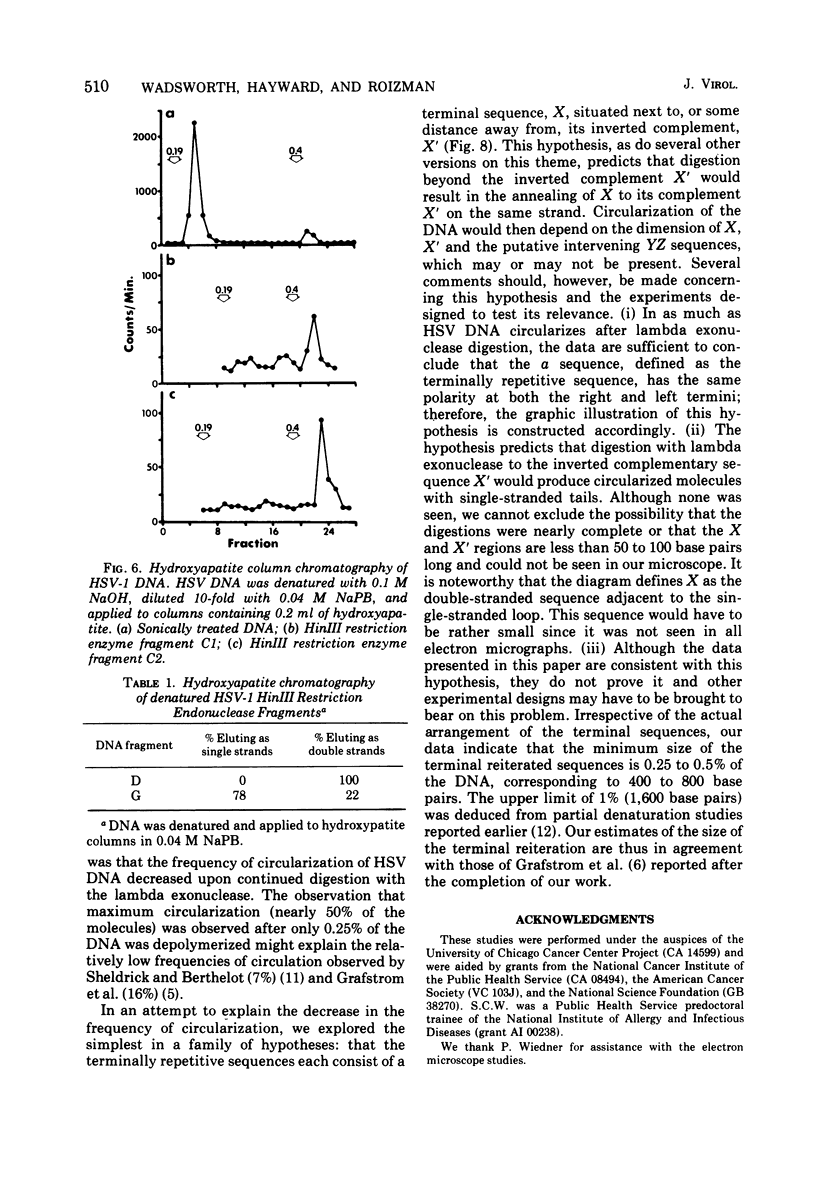
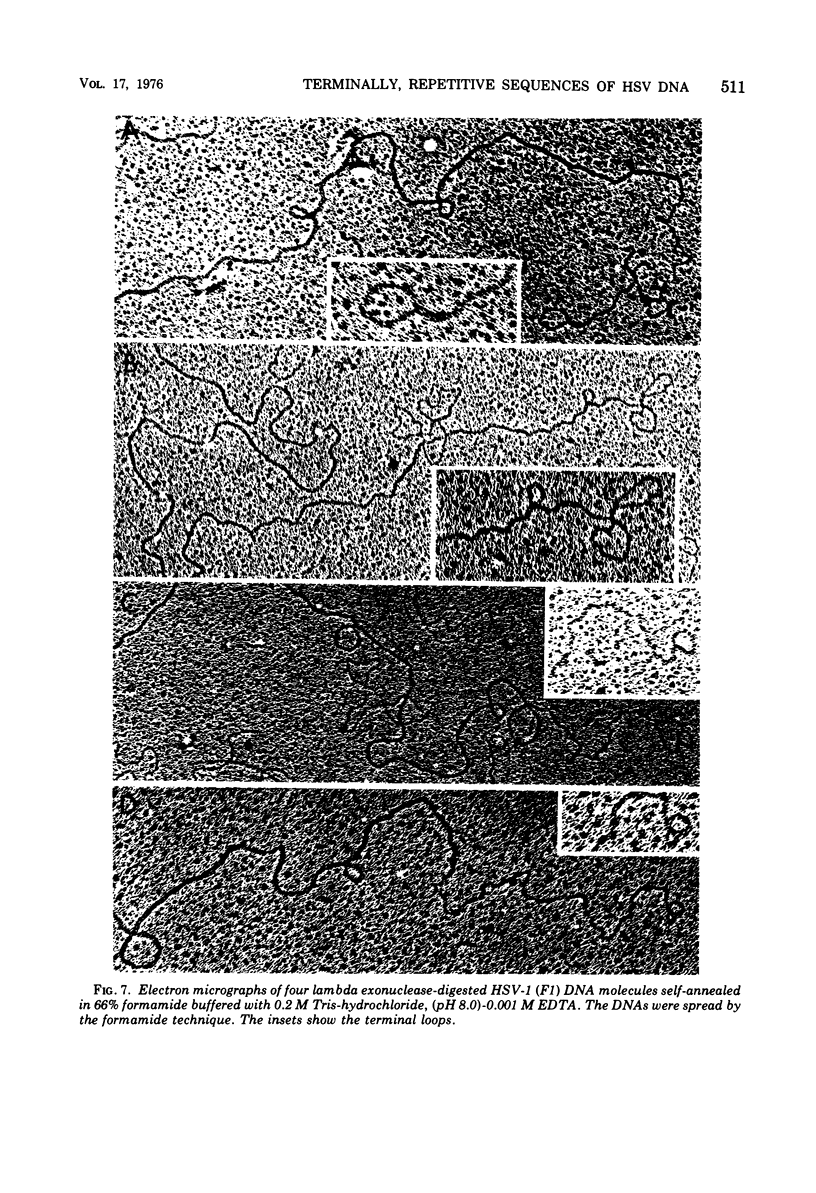
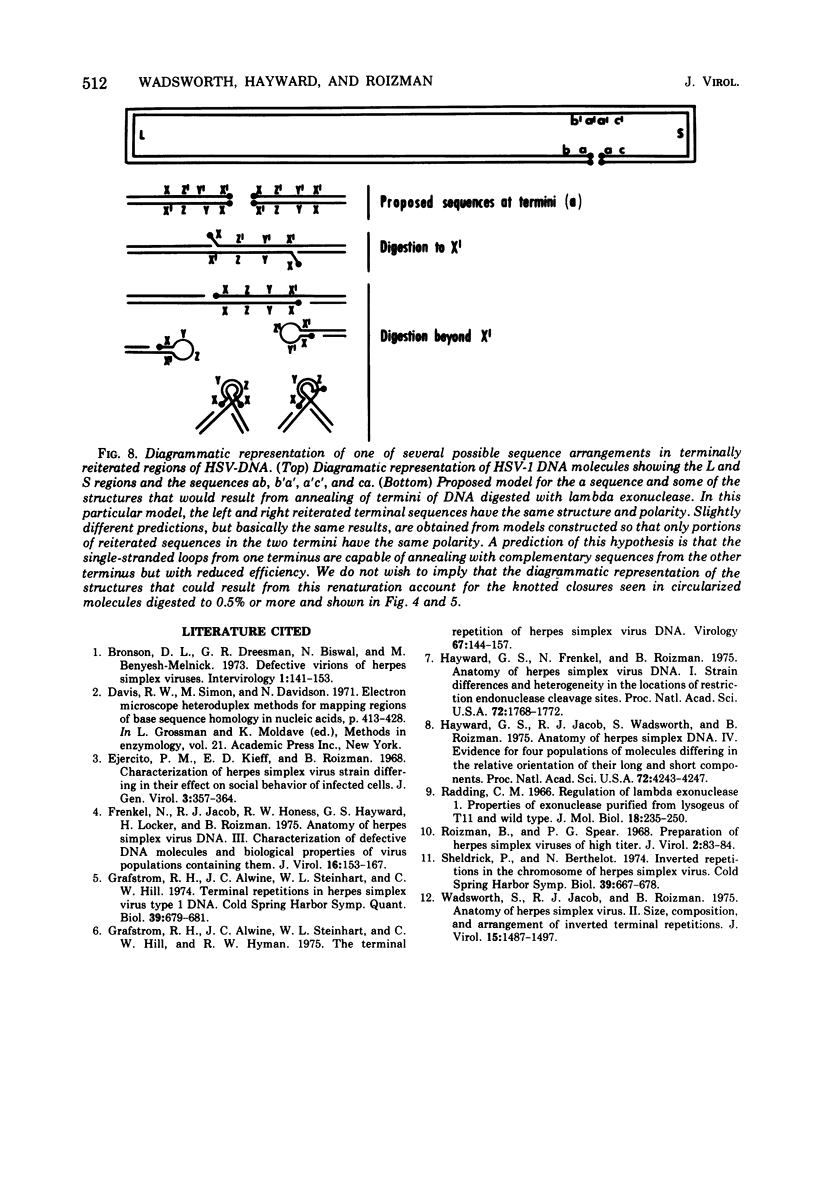
Native DNA from four strains of herpes simplex virus 1 (HSV-1) circularized after digestion with the lambda exonuclease, indicating that the molecules were terminally repetitious. In two strains, the terminal repetition was evident in nearly 50% of the DNA molecules. Maximal circularization was observed when only 0.25 to 0.5% of the DNA was depolymerized by the exonuclease, suggesting that the minimal size of the terminally repetitious regions is in the range of 400 to 800 bases pairs. More extensive exonuclease treatment resulted in a reduction in the frequency of circularization. To determine whether the terminally repetitive regions themselves contained self-annealing sequences that were precluding circularization of more extensively digested DNA, the terminal fragments from HinIII restriction endonuclease digests were isolated, denatured, and tested for their ability to self-anneal. The results of hydroxyapatite column chromatography and electron microscope examination of the terminal regions are consistent with this hypothesis.
Full text
PDF
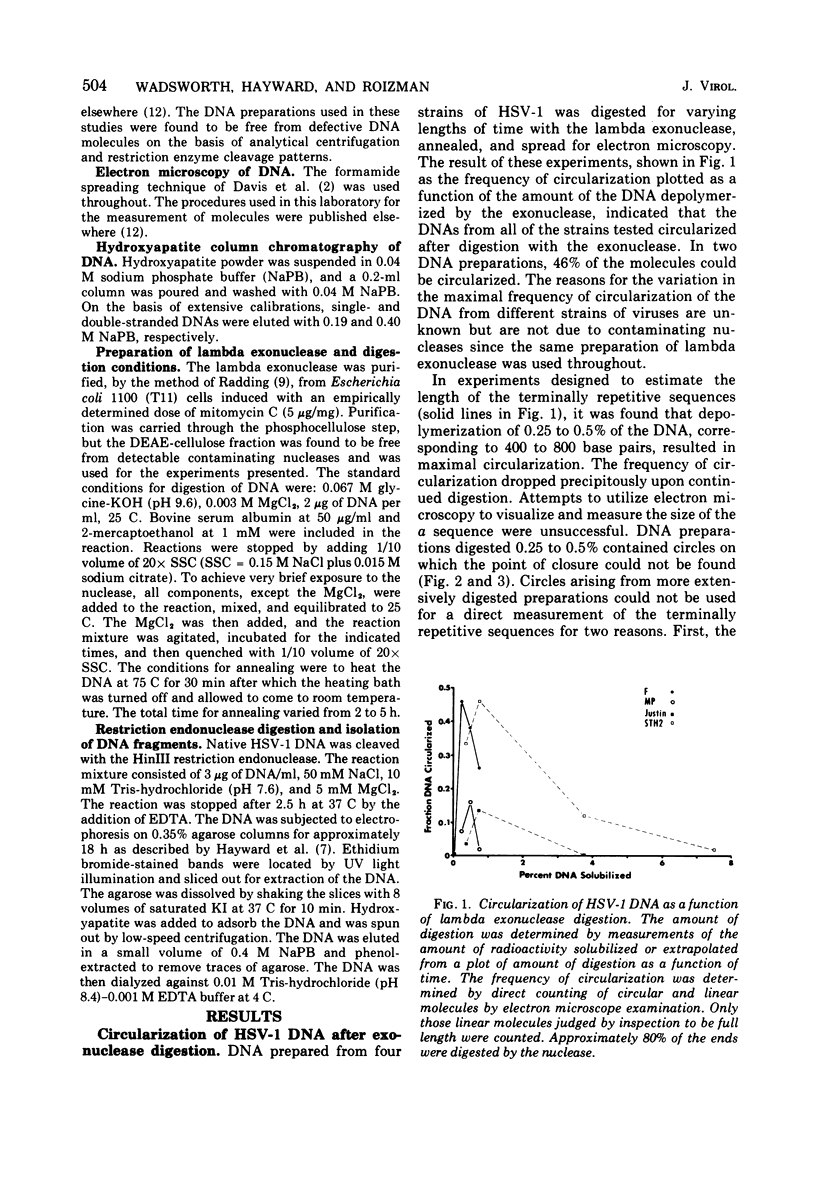

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bronson D. L., Dreesman G. R., Biswal N., Benyesh-Melnick M. Defective virions of herpes simplex viruses. Intervirology. 1973;1(3):141–153. doi: 10.1159/000148841. [DOI] [PubMed] [Google Scholar]
- Ejercito P. M., Kieff E. D., Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol. 1968 May;2(3):357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- Frenkel N., Jacob R. J., Honess R. W., Hayward G. S., Locker H., Roizman B. Anatomy of herpes simplex virus DNA. III. Characterization of defective DNA molecules and biological properties of virus populations containing them. J Virol. 1975 Jul;16(1):153–167. doi: 10.1128/jvi.16.1.153-167.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafstrom R. H., Alwine J. C., Steinhart W. L., Hill C. W., Hyman R. W. The terminal repetition of herpes simplex virus DNA. Virology. 1975 Sep;67(1):144–157. doi: 10.1016/0042-6822(75)90412-2. [DOI] [PubMed] [Google Scholar]
- Grafstrom R. H., Alwine J. C., Steinhart W. L., Hill C. W. Terminal repetitions in herpes simplex virus type 1 DNA. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):679–681. doi: 10.1101/sqb.1974.039.01.081. [DOI] [PubMed] [Google Scholar]
- Hayward G. S., Frenkel N., Roizman B. Anatomy of herpes simplex virus DNA: strain differences and heterogeneity in the locations of restriction endonuclease cleavage sites. Proc Natl Acad Sci U S A. 1975 May;72(5):1768–1772. doi: 10.1073/pnas.72.5.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward G. S., Jacob R. J., Wadsworth S. C., Roizman B. Anatomy of herpes simplex virus DNA: evidence for four populations of molecules that differ in the relative orientations of their long and short components. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4243–4247. doi: 10.1073/pnas.72.11.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radding C. M. Regulation of lambda exonuclease. I. Properties of lambda exonuclease purified from lysogens of lambda T11 and wild type. J Mol Biol. 1966 Jul;18(2):235–250. doi: 10.1016/s0022-2836(66)80243-7. [DOI] [PubMed] [Google Scholar]
- Roizman B., Spear P. G. Preparation of herpes simplex virus of high titer. J Virol. 1968 Jan;2(1):83–84. doi: 10.1128/jvi.2.1.83-84.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick P., Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Wadsworth S., Jacob R. J., Roizman B. Anatomy of herpes simplex virus DNA. II. Size, composition, and arrangement of inverted terminal repetitions. J Virol. 1975 Jun;15(6):1487–1497. doi: 10.1128/jvi.15.6.1487-1497.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]







