Abstract
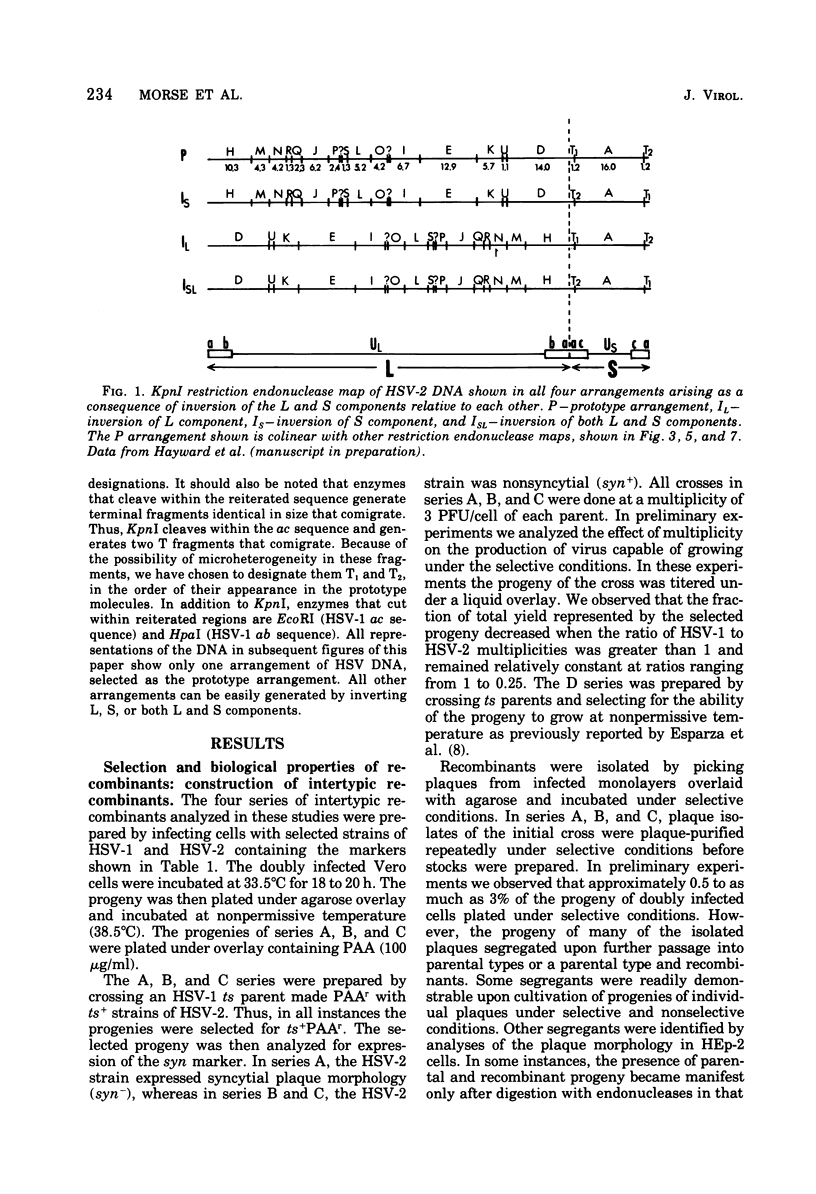
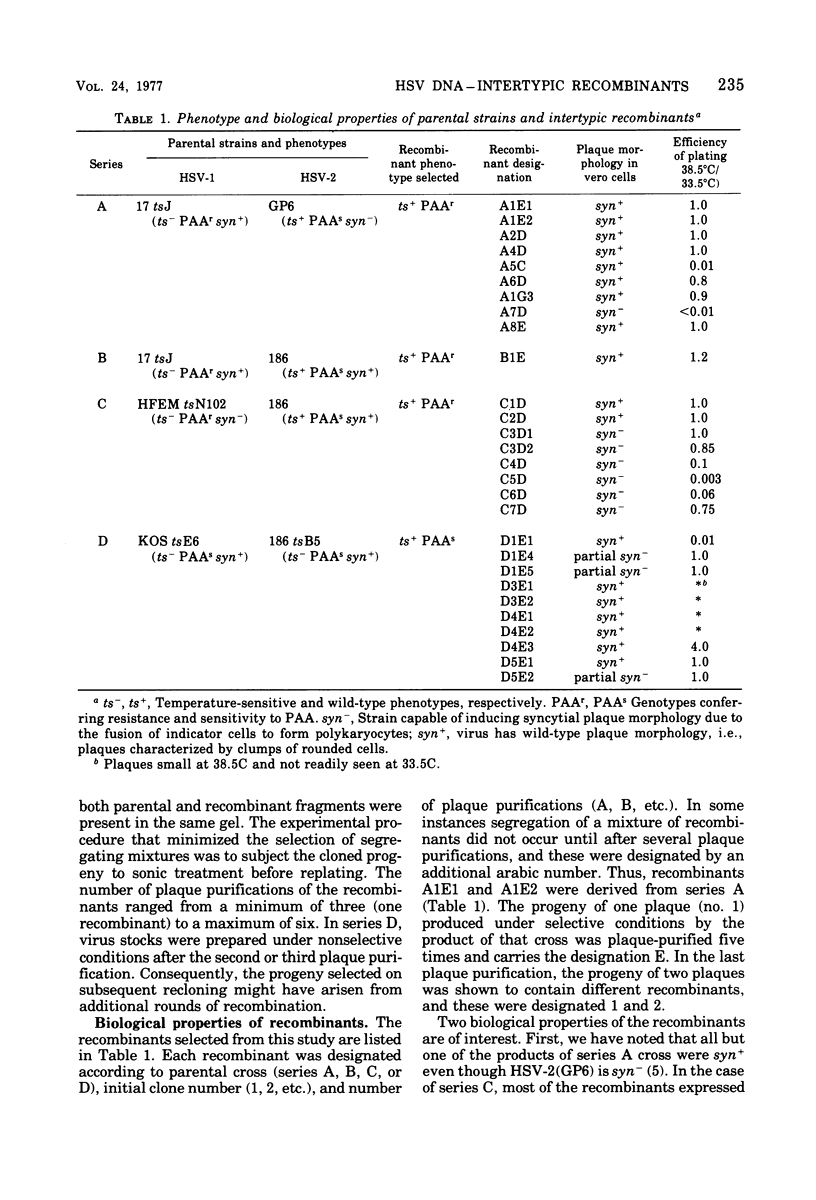
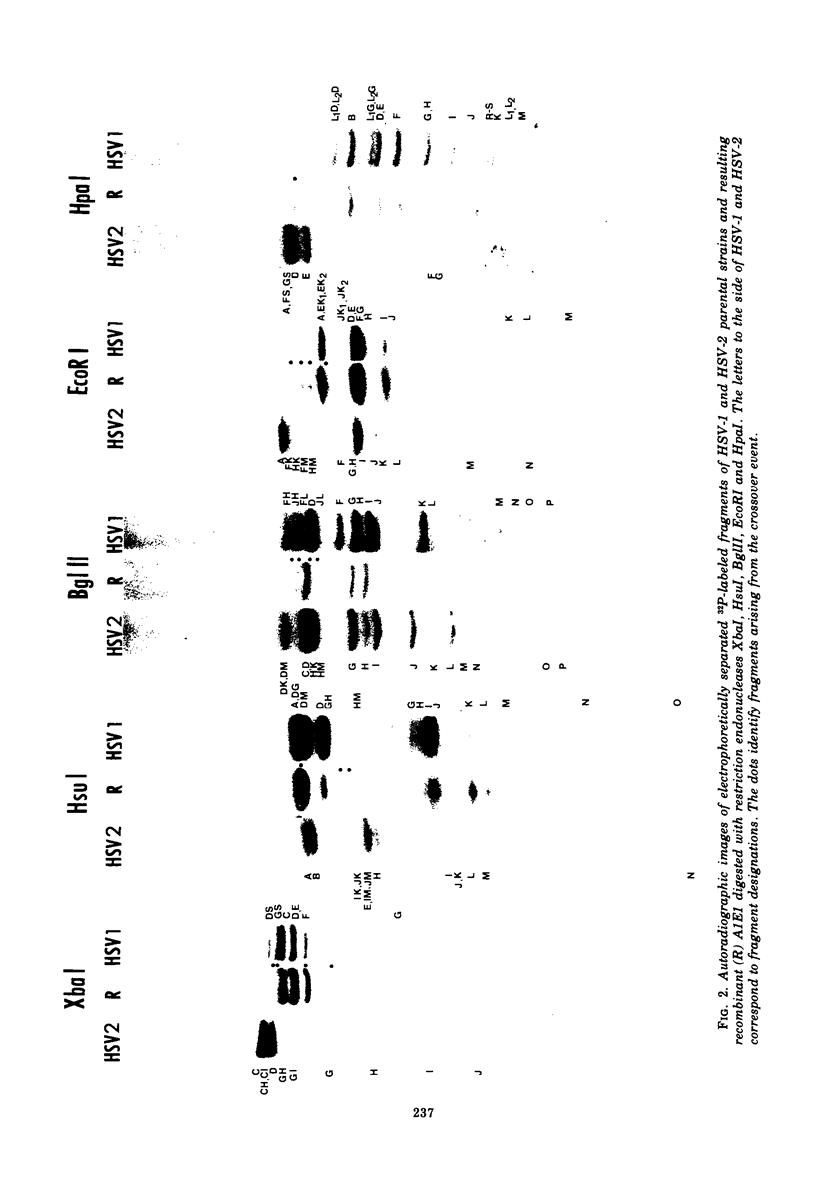
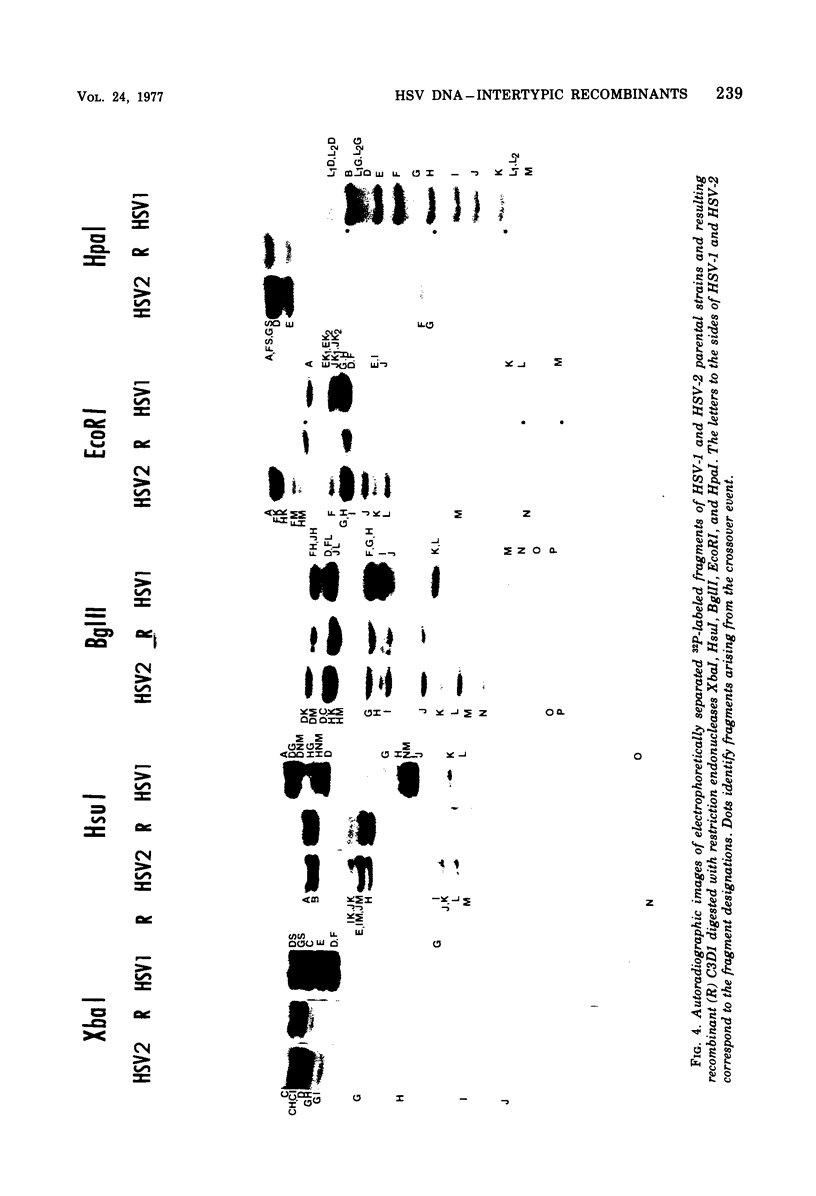
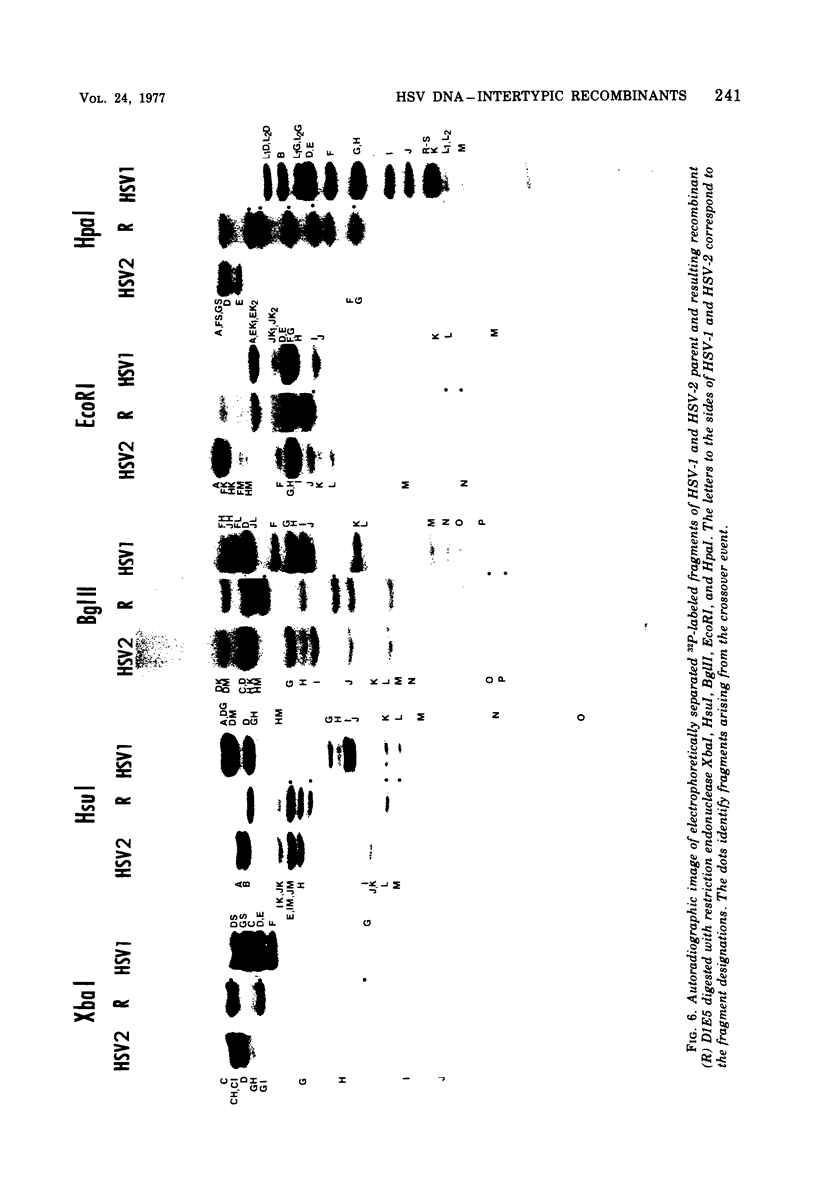
We are reporting the physical location of parental DNA sequences in 28 recombinants produced by crossing herpes simplex viruses (HSV) 1 and 2. The parental crosses were of two kinds. In the first, temperature-sensitive mutants of HSV-1 and HSV-2 were crossed to produce wild-type recombinants. In the second, temperature-sensitive mutants of HSV-1 rendered resistant to phosphonoacetic acid were crossed with wild-type HSV-2, and recombinants that multiplied at nonpermissive temperature and were resistant to the drug were selected. The DNAs of the recombinants were mapped with XbaI, EcoRI, HpaI, HsuI, BglII, and, in some instances, KpnI restriction endonucleases. The results were as follows. (i) We established the colinear arrangements of HSV-1 and HSV-2 DNAs. (ii) There was extensive interchange of genomic regions, ranging from the exchange or the entire L of S component of HSV DNA to substitutions of regions within the same component. In some recombinants, the reiterated sequences ab and ac bracketing the L and S components of HSV DNA were heterotypic. Most recombinants grew well and showed no obvious defects. (iii) The number of crossover events ranged from one to as many as six. Although crossover events occurred throughout the DNA, some clustering of crossover events was observed. (iv) Analysis of recombinants permitted localization of several markers used in this study and appears to be a useful technique for marker mapping. (v) As previously reported, HSV DNA consists of four populations, differing in relative orientation of the L and S components. All recombinants could be displayed in one arrangement of L and S such that the number of crossover events was minimized. The data are consistent with the hypothesis that only one arrangement of the parental DNA participates in the generation of recombinants.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aron G. M., Purifoy D. J., Schaffer P. A. DNA synthesis and DNA polymerase activity of herpes simplex virus type 1 temperature-sensitive mutants. J Virol. 1975 Sep;16(3):498–507. doi: 10.1128/jvi.16.3.498-507.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker Y., Dym H., Sarov I. Herpes simplex virus DNA. Virology. 1968 Oct;36(2):184–192. doi: 10.1016/0042-6822(68)90135-9. [DOI] [PubMed] [Google Scholar]
- Benyesh-Melnick M., Schaffer P. A., Courtney R. J., Esparza J., Kimura S. Viral gene functions expressed and detected by temperature-sensitive mutants of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):731–746. doi: 10.1101/sqb.1974.039.01.086. [DOI] [PubMed] [Google Scholar]
- Bronson D. L., Graham B. J., Ludwig H., Benyesh-Melnick M., Biswal N. Studies on the relatedness of herpes viruses through DNA-RNA hybridization. Biochim Biophys Acta. 1972 Jan 18;259(1):24–34. doi: 10.1016/0005-2787(72)90470-4. [DOI] [PubMed] [Google Scholar]
- Cassai E., Manservigi R., Corallini A., Terni M. Plaque dissociation of herpes simplex viruses: biochemical and biological characters of the viral variants. Intervirology. 1975;6(4-5):212–223. doi: 10.1159/000149476. [DOI] [PubMed] [Google Scholar]
- Clements J. B., Cortini R., Wilkie N. M. Analysis of herpesvirus DNA substructure by means of restriction endonucleases. J Gen Virol. 1976 Feb;30(2):243–256. doi: 10.1099/0022-1317-30-2-243. [DOI] [PubMed] [Google Scholar]
- Delius H., Clements J. B. A partial denaturation map of herpes simplex virus type 1 DNA: evidence for inversions of the unique DNA regions. J Gen Virol. 1976 Oct;33(1):125–133. doi: 10.1099/0022-1317-33-1-125. [DOI] [PubMed] [Google Scholar]
- Esparza J., Benyesh-Melnick B., Schaffer P. A. Intertypic complementation and recombination between temperature-sensitive mutants of herpes simplex virus types 1 and 2. Virology. 1976 Apr;70(2):372–384. doi: 10.1016/0042-6822(76)90279-8. [DOI] [PubMed] [Google Scholar]
- Frenkel N., Roizman B. Herpes vimplex virus: genome size and redundancy studied by renaturation kinetics. J Virol. 1971 Oct;8(4):591–593. doi: 10.1128/jvi.8.4.591-593.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkeĺ N., Locker H., Batterson W., Hayward G. S., Roizman B. Anatomy of herpes simplex virus DNA. VI. Defective DNA originates from the S component. J Virol. 1976 Nov;20(2):527–531. doi: 10.1128/jvi.20.2.527-531.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafstrom R. H., Alwine J. C., Steinhart W. L., Hill C. W., Hyman R. W. The terminal repetition of herpes simplex virus DNA. Virology. 1975 Sep;67(1):144–157. doi: 10.1016/0042-6822(75)90412-2. [DOI] [PubMed] [Google Scholar]
- Grodzicker T., Williams J., Sharp P., Sambrook J. Physical mapping of temperature-sensitive mutations of adenoviruses. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):439–446. doi: 10.1101/sqb.1974.039.01.056. [DOI] [PubMed] [Google Scholar]
- Hayward G. S., Frenkel N., Roizman B. Anatomy of herpes simplex virus DNA: strain differences and heterogeneity in the locations of restriction endonuclease cleavage sites. Proc Natl Acad Sci U S A. 1975 May;72(5):1768–1772. doi: 10.1073/pnas.72.5.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward G. S., Jacob R. J., Wadsworth S. C., Roizman B. Anatomy of herpes simplex virus DNA: evidence for four populations of molecules that differ in the relative orientations of their long and short components. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4243–4247. doi: 10.1073/pnas.72.11.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honess R. W., Roizman B. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974 Jul;14(1):8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman R. W., Burke S., Kudler L. A nearby inverted repeat of the terminal sequence of herpes simplex virus DNA. Biochem Biophys Res Commun. 1976 Jan 26;68(2):609–615. doi: 10.1016/0006-291x(76)91189-x. [DOI] [PubMed] [Google Scholar]
- Jacob R. J., Roizman B. Anatomy of herpes simplex virus DNA VIII. Properties of the replicating DNA. J Virol. 1977 Aug;23(2):394–411. doi: 10.1128/jvi.23.2.394-411.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. C., Hayward G. S., Roizman B. Anatomy of herpes simplex virus DNA VII. alpha-RNA is homologous to noncontiguous sites in both the L and S components of viral DNA. J Virol. 1977 Jan;21(1):268–276. doi: 10.1128/jvi.21.1.268-276.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieff E. D., Bachenheimer S. L., Roizman B. Size, composition, and structure of the deoxyribonucleic acid of herpes simplex virus subtypes 1 and 2. J Virol. 1971 Aug;8(2):125–132. doi: 10.1128/jvi.8.2.125-132.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieff E., Hoyer B., Bachenheimer S., Roizman B. Genetic relatedness of type 1 and type 2 herpes simplex viruses. J Virol. 1972 May;9(5):738–745. doi: 10.1128/jvi.9.5.738-745.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J. C., Robishaw E. E., Overby L. R. Inhibition of DNA polymerase from herpes simplex virus-infected wi-38 cells by phosphonoacetic Acid. J Virol. 1975 May;15(5):1281–1283. doi: 10.1128/jvi.15.5.1281-1283.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden H. S., Crombie I. K., Subak-Sharpe J. H. Control of protein synthesis in herpesvirus-infected cells: analysis of the polypeptides induced by wild type and sixteen temperature-sensitive mutants of HSV strain 17. J Gen Virol. 1976 Jun;31(3):347–372. doi: 10.1099/0022-1317-31-3-347. [DOI] [PubMed] [Google Scholar]
- Mautner V., Williams J., Sambrook J., Sharp P. A., Grodzicker T. The location of the genes coding for hexon and fiber proteins in adenovirus DNA. Cell. 1975 May;5(1):93–99. doi: 10.1016/0092-8674(75)90097-5. [DOI] [PubMed] [Google Scholar]
- Oakes J. E., Hyman R. W., Rapp F. Genome location of polyadenylated transcripts of herpes simplex virus type 1 and type 2 DNA. Virology. 1976 Nov;75(1):145–154. doi: 10.1016/0042-6822(76)90013-1. [DOI] [PubMed] [Google Scholar]
- Overby L. R., Robishaw E. E., Schleicher J. B., Rueter A., Shipkowitz N. L., Mao J. C. Inhibition of herpes simplex virus replication by phosphonoacetic acid. Antimicrob Agents Chemother. 1974 Sep;6(3):360–365. doi: 10.1128/aac.6.3.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purifoy D. J., Benyesh-Melnick M. DNA polymerase induction by DNA-negative temperature-sensitive mutants of herpes simplex virus type 2. Virology. 1975 Dec;68(2):374–386. doi: 10.1016/0042-6822(75)90280-9. [DOI] [PubMed] [Google Scholar]
- ROIZMAN B., ROANE P. R., Jr A physical difference between two strains of herpes simplex virus apparent on sedimentation in cesium chloride. Virology. 1961 Sep;15:75–79. doi: 10.1016/0042-6822(61)90079-4. [DOI] [PubMed] [Google Scholar]
- Roizman B., Hayward G., Jacob R., Wadsworth S., Frenkel N., Honess R. W., Kozak M. Human herpersviruses I: a model for molecular organization and regulation of herpesviruses-a review. IARC Sci Publ. 1975;(11 Pt 1):3–38. [PubMed] [Google Scholar]
- Roizman B., Kozak M., Honess R. W., Hayward G. Regulation of herpesvirus macromolecular synthesis: evidence for multilevel regulation of herpes simplex 1 RNA and protein synthesis. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):687–701. doi: 10.1101/sqb.1974.039.01.083. [DOI] [PubMed] [Google Scholar]
- Schaffer P. A. Temperature-sensitive mutants of herpesviruses. Curr Top Microbiol Immunol. 1975;70:51–100. doi: 10.1007/978-3-642-66101-3_3. [DOI] [PubMed] [Google Scholar]
- Sheldrick P., Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Skare J., Summers W. C. Structure and function of herpesvirus genomes. II. EcoRl, Sbal, and HindIII endonuclease cleavage sites on herpes simplex virus. Virology. 1977 Feb;76(2):581–595. doi: 10.1016/0042-6822(77)90240-9. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Nathans D. Letter: A suggested nomenclature for bacterial host modification and restriction systems and their enzymes. J Mol Biol. 1973 Dec 15;81(3):419–423. doi: 10.1016/0022-2836(73)90152-6. [DOI] [PubMed] [Google Scholar]
- Subak-Sharpe J. H., Brown S. M., Ritchie D. A., Timbury M. C., Macnab J. C., Marsden H. S., Hay J. Genetic and biochemical studies with herpesvirus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):717–730. doi: 10.1101/sqb.1974.039.01.085. [DOI] [PubMed] [Google Scholar]
- Wadsworth S., Hayward G. S., Roizman B. Anatomy of herpes simplex virus DNA. V. Terminally repetitive sequences. J Virol. 1976 Feb;17(2):503–512. doi: 10.1128/jvi.17.2.503-512.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth S., Jacob R. J., Roizman B. Anatomy of herpes simplex virus DNA. II. Size, composition, and arrangement of inverted terminal repetitions. J Virol. 1975 Jun;15(6):1487–1497. doi: 10.1128/jvi.15.6.1487-1497.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie N. M., Clements J. B., Macnab J. C., Subak-Sharpe J. H. The structure and biological properties of herpes simplex virus DNA. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):657–666. doi: 10.1101/sqb.1974.039.01.079. [DOI] [PubMed] [Google Scholar]
- Wilkie N. M., Cortini R. Sequence arrangement in herpes simplex virus type 1 DNA: identification of terminal fragments in restriction endonuclease digests and evidence for inversions in redundant and unique sequences. J Virol. 1976 Oct;20(1):211–221. doi: 10.1128/jvi.20.1.211-221.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie N. M. Physical maps for Herpes simplex virus type 1 DNA for restriction endonucleases Hind III, Hpa-1, and X. bad. J Virol. 1976 Oct;20(1):222–233. doi: 10.1128/jvi.20.1.222-233.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]