Abstract
DNA can be sequenced by a chemical procedure that breaks a terminally labeled DNA molecule partially at each repetition of a base. The lengths of the labeled fragments then identify the positions of that base. We describe reactions that cleave DNA preferentially at guanines, at adenines, at cytosines and thymines equally, and at cytosines alone. When the products of these four reactions are resolved by size, by electrophoresis on a polyacrylamide gel, the DNA sequence can be read from the pattern of radioactive bands. The technique will permit sequencing of at least 100 bases from the point of labeling.
Full text
PDF


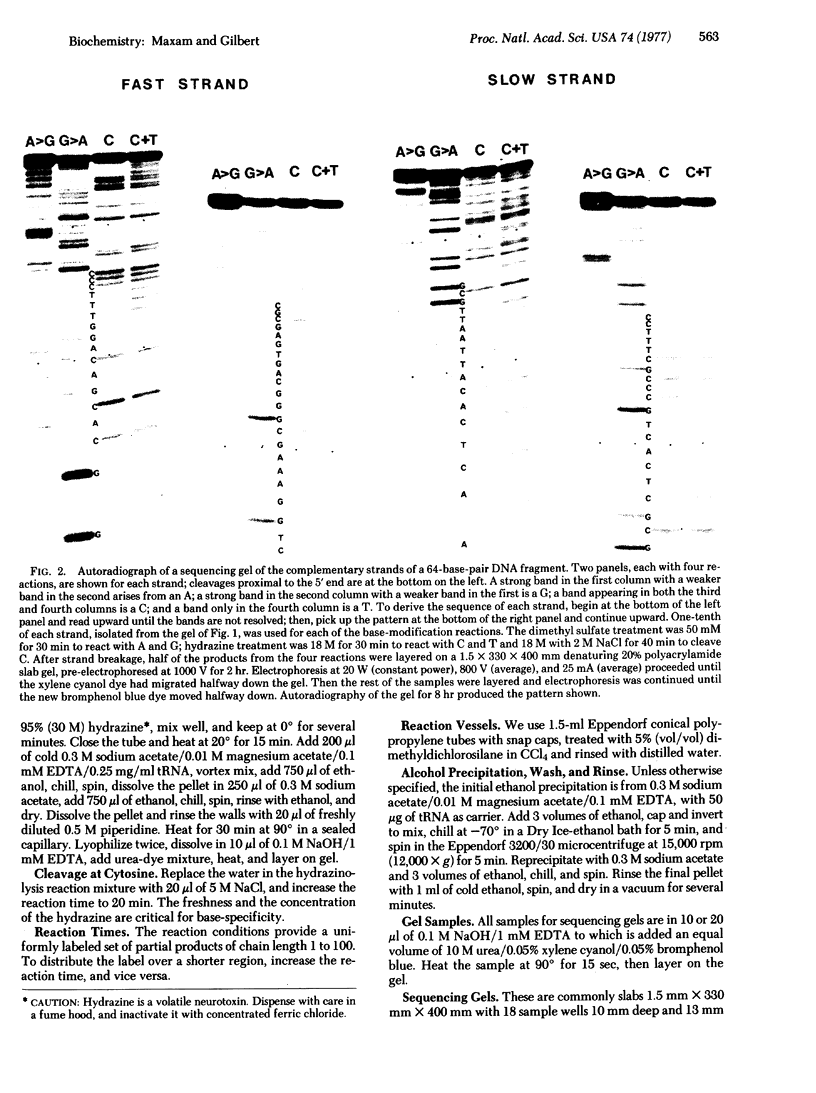

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cashmore A. R., Petersen G. B. The degradation of DNA by hydrazine: a critical study of the suitability of the reaction for the quantitative determination of purine nucleotide sequences. Biochim Biophys Acta. 1969 Feb 18;174(2):591–603. doi: 10.1016/0005-2787(69)90289-5. [DOI] [PubMed] [Google Scholar]
- Glynn I. M., Chappell J. B. A simple method for the preparation of 32-P-labelled adenosine triphosphate of high specific activity. Biochem J. 1964 Jan;90(1):147–149. doi: 10.1042/bj0900147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes D. H., Hayes-Baron F. Hydrazinolysis of some purines and pyrimidines and their related nucleosides and nucleotides. J Chem Soc Perkin 1. 1967;16:1528–1533. doi: 10.1039/j39670001528. [DOI] [PubMed] [Google Scholar]
- Hayward G. S. Gel electrophoretic separation of the complementary strands of bacteriophage DNA. Virology. 1972 Jul;49(1):342–344. doi: 10.1016/s0042-6822(72)80042-4. [DOI] [PubMed] [Google Scholar]
- KRIEK E., EMMELOT P. METHYLATION OF DEOXYRIBONUCLEIC ACID BY DIAZOMETHANE. Biochim Biophys Acta. 1964 Sep 11;91:59–66. doi: 10.1016/0926-6550(64)90170-7. [DOI] [PubMed] [Google Scholar]
- LAWLEY P. D., BROOKES P. FURTHER STUDIES ON THE ALKYLATION OF NUCLEIC ACIDS AND THEIR CONSTITUENT NUCLEOTIDES. Biochem J. 1963 Oct;89:127–138. doi: 10.1042/bj0890127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillehaug J. R., Kleppe K. Effect of salts and polyamines on T4 polynucleotide kinase. Biochemistry. 1975 Mar 25;14(6):1225–1229. doi: 10.1021/bi00677a021. [DOI] [PubMed] [Google Scholar]
- Roberts R. J., Myers P. A., Morrison A., Murray K. A specific endonuclease from Arthrobacter luteus. J Mol Biol. 1976 Mar 25;102(1):157–165. doi: 10.1016/0022-2836(76)90079-6. [DOI] [PubMed] [Google Scholar]
- Roychoudhury R., Jay E., Wu R. Terminal labeling and addition of homopolymer tracts to duplex DNA fragments by terminal deoxynucleotidyl transferase. Nucleic Acids Res. 1976 Apr;3(4):863–877. doi: 10.1093/nar/3.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TEMPERLI A., TUERLER H., RUEST P., DANON A., CHARGAFF E. STUDIES OF THE NUCLEOTIDE ARRANGEMENT IN DEOXYRIBONUCLEIC ACIDS. IX. SELECTIVE DEGRADATION OF PYRIMIDINE DEOXYRIBONUCLEOTIDES. Biochim Biophys Acta. 1964 Nov 15;91:462–476. doi: 10.1016/0926-6550(64)90076-3. [DOI] [PubMed] [Google Scholar]
- van de Sande J. H., Kleppe K., Khorana H. G. Reversal of bacteriophage T4 induced polynucleotide kinase action. Biochemistry. 1973 Dec 4;12(25):5050–5055. doi: 10.1021/bi00749a004. [DOI] [PubMed] [Google Scholar]